Antarctic ice algae CPD photolyase, and coding gene, expression vector and application thereof
A technology of photorepair enzymes and Antarctic ice algae, applied in the field of biology, can solve the problems of research difficulties such as the separation and purification of biological photorepair enzymes and in vitro activity, and the lack of photorepair enzymes. benefit effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
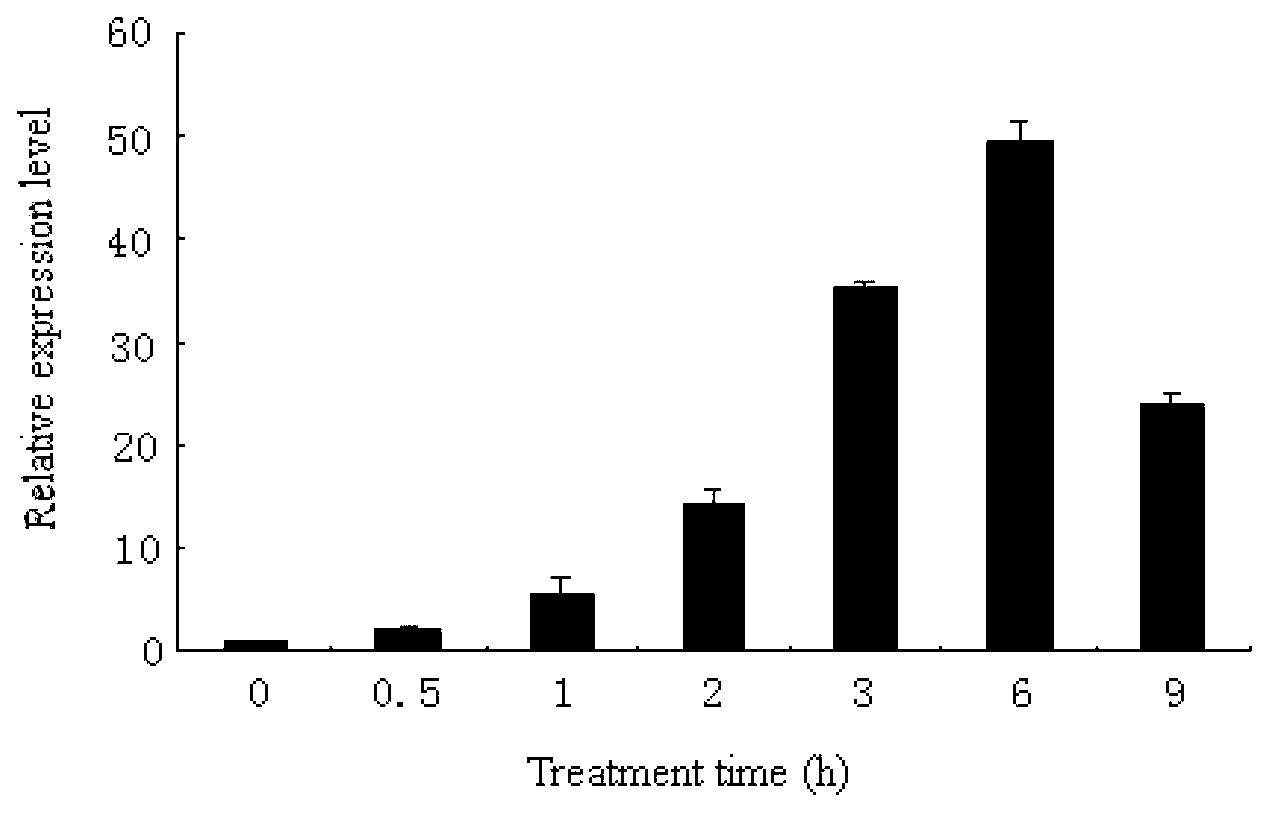
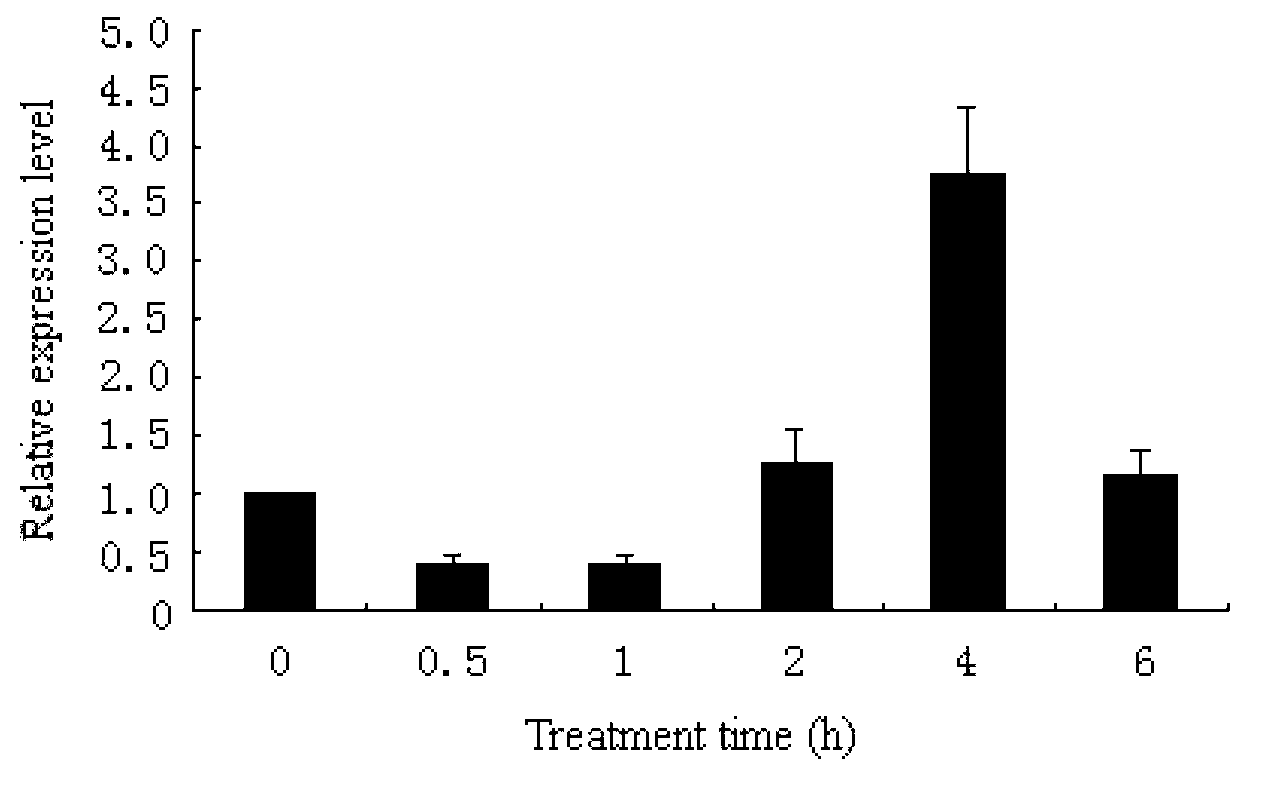
[0041] Example 1. Obtaining the cDNA sequence of the Antarctic ice alga Chlamydomonas sp.ICE-L CPD photorepair enzyme
[0042] 1. Cultivation of Antarctic ice algae and isolation of total RNA
[0043] The Antarctic ice algae were cultured to the logarithmic phase at 4°C, and the RNA was extracted with Trizol from Invitrogen Company. The test was carried out according to the extraction process instructions of Trizol reagent. No RNase contamination was ensured throughout the process. The extracted RNA was divided into small portions and stored in a -80°C refrigerator for later use.
[0044] 2. Acquisition of cDNA sequence of CPD photorepair enzyme gene
[0045] Primers were designed according to the ice algae transcriptome sequence obtained earlier:
[0046] 5'-RACE Primer GSP1:CGAGCACATGGGTCGTTCCCTCTTATCG
[0047] 3'-RACE Primer GSP2:TGGGACGGAAGTGGAGGGATGAGGT
[0048] The obtained high-quality RNA was reverse-transcribed to synthesize cDNA.
[0049] The 5'-RACE reverse tra...
Embodiment 2
[0057] Example 2. Construction of Antarctic ice alga Chlamydomonas sp.ICE-L CPD photorepair enzyme expression vector
[0058] 1. Design primers according to the complete sequence of the photorepair enzyme gene obtained by sequencing:
[0059] CPDF:TAGAATTCATGCCGAAGCGAAC
[0060] CPDR: TATCTCGAGTCAAGCTCGTGGC
[0061] The PCR reaction conditions were: pre-denaturation at 95°C for 10 minutes, denaturation at 95°C for 15 seconds, annealing at 55°C for 1 minute, and extension at 72°C for 2 minutes for 30 cycles.
[0062] 2. The PCR amplification product of the photorepair enzyme gene and the prokaryotic expression vector pET-28a(+) were respectively digested with EcoRI and XhoI at 37°C for 2 hours, recovered with 1.0% agarose, and the purified target fragment And the vector was ligated with T4 DNA ligase at 16°C for 16h, and the ligation system was as follows.
[0063]
[0064] 3. Transform the ligated recombinant plasmid into Escherichia coli DH5α, the steps are as follows: ...
Embodiment 3
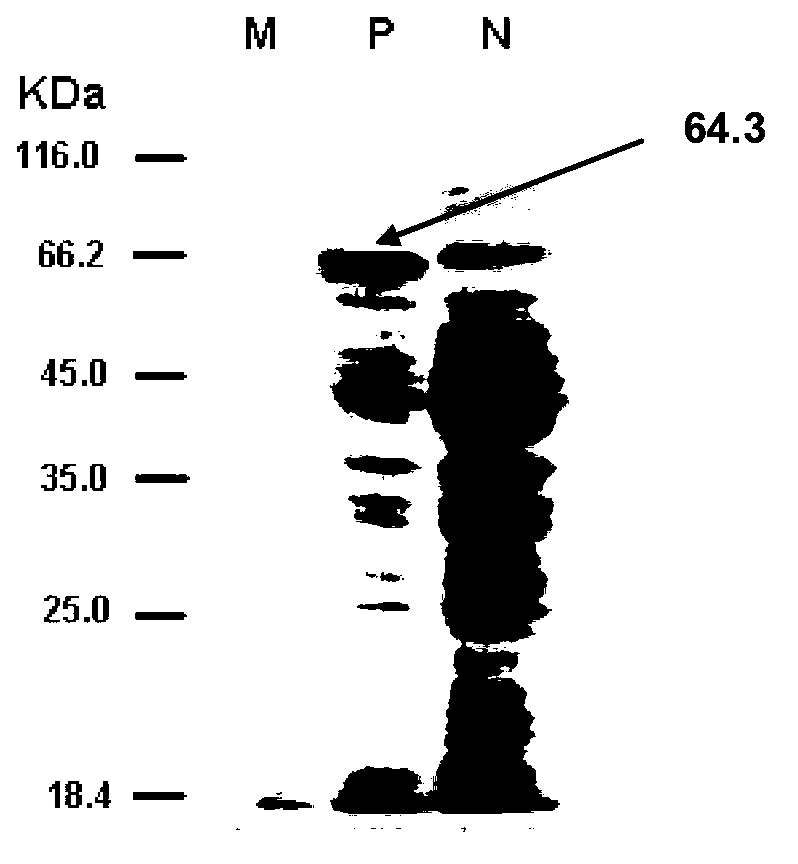
[0072] Example 3. Purification and protein detection of Antarctic ice algae Chlamydomonas sp.ICE-L CPD photorepair enzyme
[0073] 1. Inoculate the recombinant bacterium BL21(DE3) / pET-28a(+) / phr in 5 mL of LB liquid medium containing kanamycin, and cultivate overnight at 37°C with shaking. On the next day, take 5 mL of the bacterial liquid and insert it into 500 mL of the same medium to continue culturing for 2 to 3 hours (OD600 about 0.6), then add IPTG at a final concentration of 1 mM, shake and culture at 10°C for 24 hours, centrifuge at 8000 g for 10 minutes, and collect the bacteria.
[0074] 2. Ni agarose affinity column affinity chromatography: put the metal chelating agent Ni agarose into the chromatographic column, and equilibrate with 1×Ni column binding buffer. Apply the supernatant to the column. Generally, 40mM imidazole is used to elute to obtain the target protein.
[0075] 3. Take the above induced BL21(DE3) / pET-28a(+) / phr cells and the purified protein expre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sub-mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com