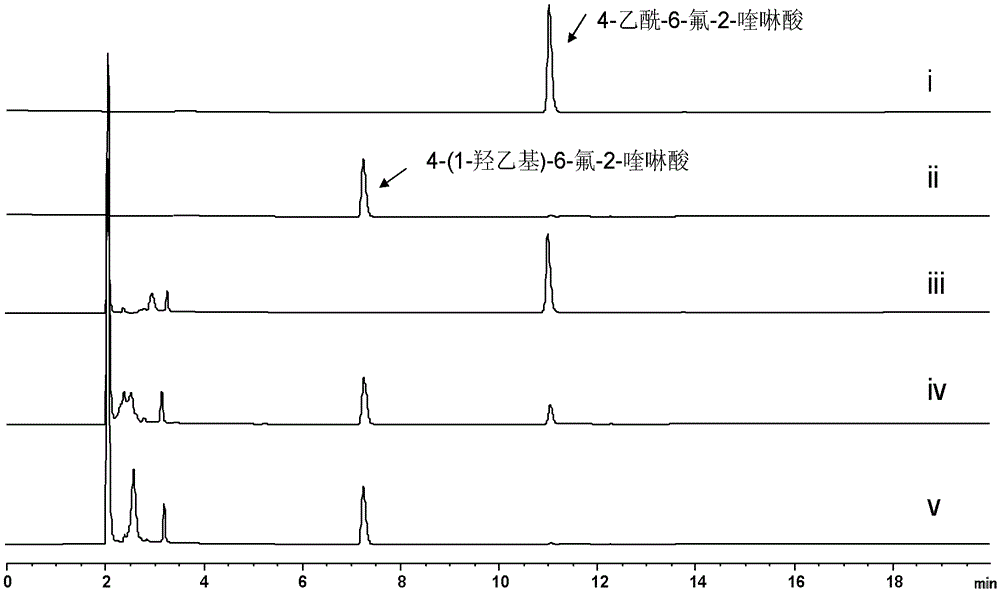
Function and application of a short-chain dehydrogenase tsru
A short-chain dehydrogenase, a technology for uses, applied in the fields of biotechnology and engineering, and can solve problems such as complex structures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0093] In a preferred example of the present invention, the preparation method of TsrU comprises steps:
[0094] (1) Obtain a 0.85kb DNA fragment encoding the short-chain dehydrogenase TsrU from the Streptomyceslaurentii genome by PCR, then connect it to a commercially available plasmid vector, and introduce the successfully constructed plasmid into an E. coli host to obtain a recombinant strain. Induce the expression of host TsrU;
[0095] (2) Centrifuge the cultured bacterial strains, collect the bacterial cells, resuspend the bacterial cells in a bacteriostasis buffer (50 mM Tris-Cl, 200 mM NaCl, pH=8.0), and sonicate the bacterium. After breaking the bacteria, the suspended liquid was centrifuged, and the supernatant was combined with Ni-NTA overnight, washed with the bacteria breaking buffer containing different concentrations of imidazole, and the protein TsrU was obtained in the washing solution of the imidazole solution. TsrU can be stored at -80°C for a long time.
...
Embodiment 1
[0137] Construction of Escherichia coli Recombinant Strain SL1121
[0138] The sequence of primers for cloning the gene encoding the TsrU protein is as follows:
[0139] TsrU-F (SEQ ID NO: 3) 5'-CGAAGCTTCAGGTGGTGCCGTCAGACG-3';
[0140] TsrU-R (SEQ ID NO: 4) 5'-CATATGACCGCCCCCGCGCTCCCGCTC-3'.
[0141] Using the total DNA of Streptomyceslaurentii ACTCC31255, a thiostrepton-producing bacterium, as a template, a PCR reaction system was composed of dNTP, DMSO, enzyme-free water, high-fidelity Primestar DNA polymerase and its buffer to express TsrU gene The 0.85kb fragment was amplified by PCR. After the amplification reaction, the reaction product was separated by gel electrophoresis, recovered and purified by cutting the gel, after purification, restriction endonuclease HindIII and NdeI were added to digest and recover the fragment, and it was connected into pET28a treated with the same enzyme to construct a recombinant vector . The vector was transformed into E.coliDH5α, and ...
Embodiment 2
[0143] Heterologous expression of short-chain dehydrogenase TsrU
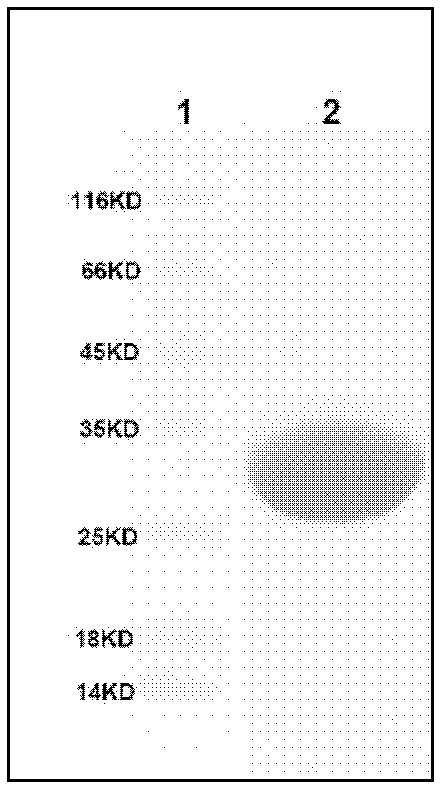
[0144] Inoculate Escherichia coli SL1121 in 3ml LB medium (Kanamycin 50μg / ml) and incubate at 37°C for 8 hours, then inoculate in 800ml LB medium (Kanamycin 50μg / ml), and incubate at 37°C for 2.5 hours to A 600nm 0.5-0.7. Then transfer to 25° C. to continue culturing for 0.5 hours, add IPTG to a final concentration of 0.1 mM, and culture at 25° C. for more than 6 hours (or overnight). The bacterial liquid was centrifuged (5000 rpm, 20 minutes), and the bacterial cells were collected. The bacteria were disrupted by ultrasonic waves, the broken bacteria were centrifuged (15000 rpm, 30 minutes), and the supernatant solution was mixed with 3 mL of Ni-NTA (QIAGEN) overnight. Finally, wash with imidazole solutions with gradient concentrations of 100, 150, 200, 250, and 300 mM respectively, collect eluate from 150 to 200 mM, concentrate and desalt, and obtain purified TsrU protein.
[0145] figure 2 The SDS-PAGE ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com