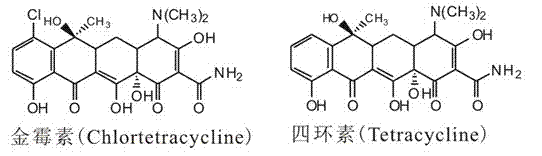
Method for improving aureomycin yield as well as recombinant expression vector and genetic engineering bacterium of aureomycin
A technology of genetically engineered bacteria and expression vectors, applied in the directions of microorganism-based methods, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of limited production capacity of chlortetracycline, and achieve the goal of improving the production of chlortetracycline and improving the Yield, simple effect of preparation method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
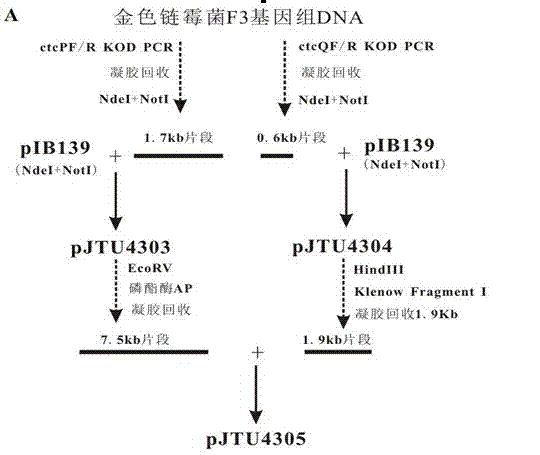
[0034] construct with FADH 2 dependent chlorase gene ctcP and FAD reductase gene ctQ A recombinant expression plasmid, specifically an integrative plasmid.
[0035] According to FADH 2 dependent chlorase gene ctcPSequence (Genebank: I32939) Design Amplification ctcP The PCR primers of the gene, and the NdeI restriction site was introduced at the 5' end of the upstream primer, and the NotI was introduced at the 5' end of the downstream primer. The specific primers are as follows:
[0036] Upstream primer ctcPF: 5'-ggaattc catatg accgacacaaccgc-3' (SEQ ID NO.1), the underlined sequence is the NdeI site, and the boldface is the start codon;
[0037] Downstream primer ctcPR: 5'- gcggccgc aagcttctacggcttgacccgcatcg-3' (SEQ ID NO.2), the underlined sequence is the NotI site, and the boldface is the stop codon.
[0038] Then, Streptomyces aureus F3 genomic DNA was used as a template, and the sequences shown in SEQ ID NO.1 and SEQ ID NO.2 were used as primers for PCR amp...
Embodiment 2
[0045] The constructed integrated plasmid pJTU4305 was transformed into Streptomyces aureus F3 by conjugative transfer between Escherichia coli and Streptomyces parents. The specific steps include: transforming the constructed integrated plasmid pJTU4305 into Escherichia coli ET12567 (pUZ8002) by heat shock at 42°C, selecting a single clone transformant to inoculate LB liquid medium, adding a final concentration of 25 μg / mL to the liquid medium Chloramphenicol, 50 μg / mL kanamycin and 30 μg / mL abramycin (Escherichia coli ET12567 (pUZ8002) Published in Paget, M.S., Chamberlin, L., Atrih, A., Paget, M.S. et.al . Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3 (2). J. Bacteriol, 1999, 181: 204-211.). Then culture the positive clones overnight at 37°C, transfer the inoculum to 5 mL of fresh LB liquid medium containing the same antibiotic according to the volume fraction of 10%, collect the bacter...
Embodiment 3
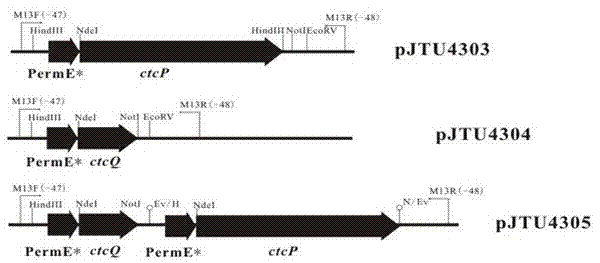
[0047] Pick a single clone of the binder, mark a single colony on LB solid medium containing 30 μg / mL abramycin, and culture it in a 30°C incubator. After relaxation on the medium, spores were collected with 20% glycerol after 6-7 days of culture at 30°C. Spores use 10 -4 、10 -6 、10 -8 、10 -10 The plate containing abramycin was diluted in a concentration gradient, cultured at 30°C for 3 days, and a plate with a suitable number of single colonies was selected to inoculate 10.3% YEME liquid medium, and then genomic DNA was extracted for PCR verification. The sequence of the verification primers was as follows:
[0048] Upstream primer M13F(-47): 5'-cgccagggttttcccagtcacgac-3' (SEQ ID NO.7);
[0049] Downstream primer M13R(-48): 5'-agcggataacaatttcacacagga-3' (SEQ ID NO.8).
[0050] Test results such as Figure 4 As shown, a positive clone was obtained and named Streptomyces aureus ZT03. Streptomyces aureus ZT03 is preserved with the oat plate that contains 20% glycerol by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com