Kit for detecting salmonella in foods and application method thereof
A Salmonella and kit technology, applied in the direction of biochemical equipment and methods, microbe measurement/inspection, and resistance to vector-borne diseases, etc., can solve the problems of long time consumption and low sensitivity, and achieve simple reaction steps, low cost, and detection short-term effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 The kit of the present invention
[0041] The kit: includes 24 reaction tubes, each containing 23μL of reaction mixture and 30μL of liquid paraffin; 1 tube of positive control (50μL); 2 tubes of DNA extraction reagent A (1300μL / tube); 1 tube of DNA extraction Reagent B (600μL). The reaction mixture contains: outer primer F3 with a concentration of 0.2 μM, the nucleotide sequence of which is shown in SEQ ID NO: 1; and outer primer B3 with a concentration of 0.2 μM, whose nucleotide sequence is as shown in SEQ ID NO: 2 The nucleotide sequence of the inner primer FIP with a concentration of 1.6 μM is shown in SEQ ID NO: 3; the nucleotide sequence of the inner primer BIP with a concentration of 1.6 μM is shown in SEQ ID NO: 4; The nucleotide sequence of the loop primer LoopF of 0.8μM is shown in SEQ ID NO:5; the nucleotide sequence of the loop primer LoopB of 0.8μM is shown in SEQ ID NO:6; 20mM pH8.8 Tris buffer, 10mM KCl, 10mM NH 4 SO 4 , 0.1% by weight of Tween...
Embodiment 2
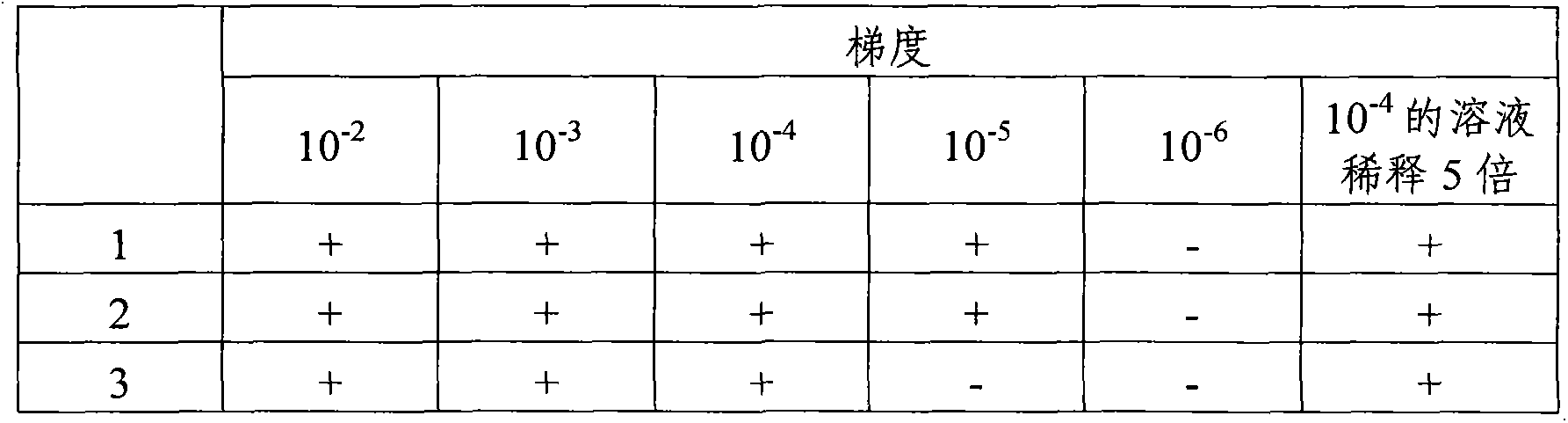
[0043] Example 2 Sensitivity test of the method of the present invention
[0044] Preparation of BPW medium: Weigh 4.5 g of CM201 buffered peptone water medium (purchased from Beijing Luqiao Technology Co., Ltd.), add water to prepare 225 mL of enrichment medium, which is called BPW medium. The pH value is 7.0±0.2. Sterilize at 121°C for 15 minutes, and then set aside.
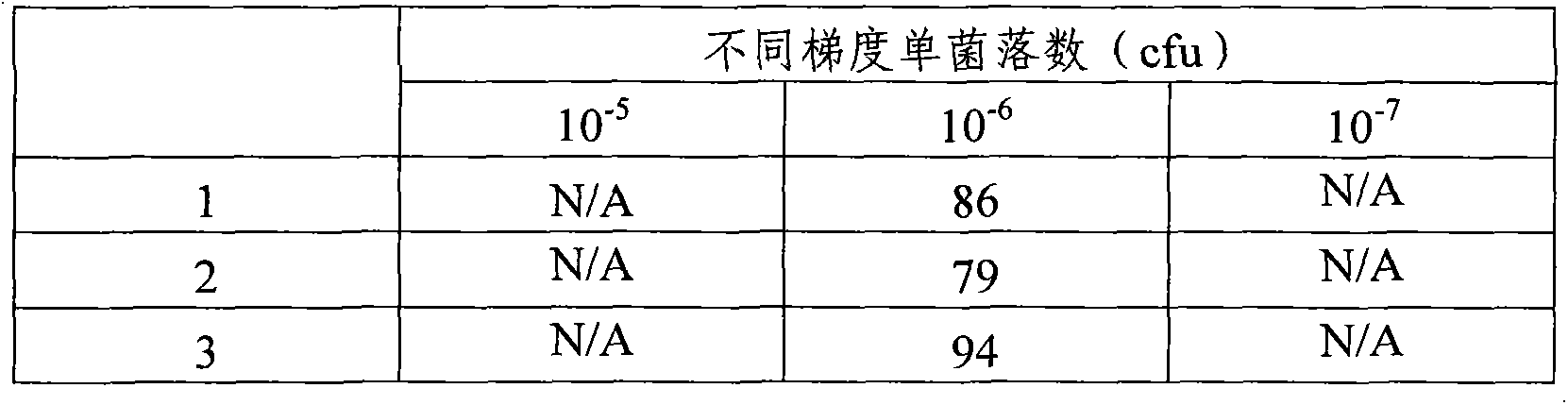
[0045] Salmonella was inoculated into 4 mL of BPW medium and cultured overnight at 37°C. 40 μL of overnight culture broth was inoculated into another new 4 mL BPW medium and cultured at 35°C at 150 rpm for 4 hours to obtain metaphase cells. Then it was diluted with normal saline 10-fold gradient dilution method, and the treatment of each concentration was performed three times. Select 10 -2 , 10 -3 , 10 -4 , 10 -5 , 10 -6 , 10 -7 100 μL of solutions of 6 dilutions were spread on TSA plates (purchased from Beijing Luqiao Technology Co., Ltd.) and incubated at 37°C overnight. Colony count results are shown in Tabl...
Embodiment 3
[0053] Example 3 Specificity test of the method of the present invention
[0054] Pick 13 strains of Salmonella (Sal), 6 strains of Escherichia coli (E.coli), 8 strains of Vibrio parahaemolyticus (VP), 2 strains of Shigella (Shi), and 12 strains of Listeria monocytogenes. The bacteria (LM) and 6 strains of Staphylococcus aureus (SA) were added to 200μL of sterilized deionized water, heated at 98°C for 10min, centrifuged at 7000g for 2min, and the supernatant was taken as the LAMP detection DNA template solution.
[0055] Add 2 μL of the DNA template solution prepared above into a reaction tube containing 23 μL of the reaction solution described in Example 1 and 30 μL of liquid paraffin, and react at 64° C. for 60 min. The positive is green and the negative is orange.
[0056] The results showed that only the Salmonella test tubes were positive, and the test tubes for other strains were negative.
[0057]
[0058]
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com