Long double-stranded RNA expression vector for silencing whole genome or across genome
An expression vector and genome-wide technology, applied in the fields of RNA interference technology and functional genomics, can solve the problems of immature high-throughput RNA interference vectors and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
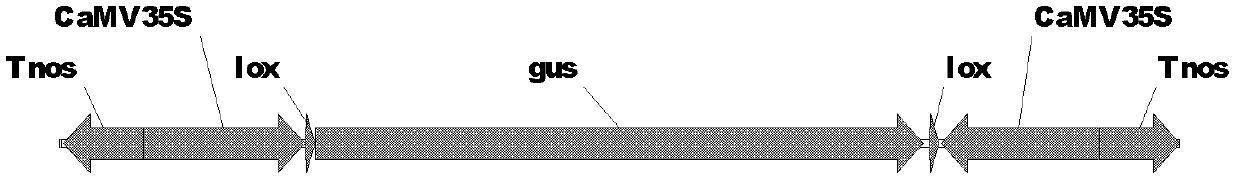
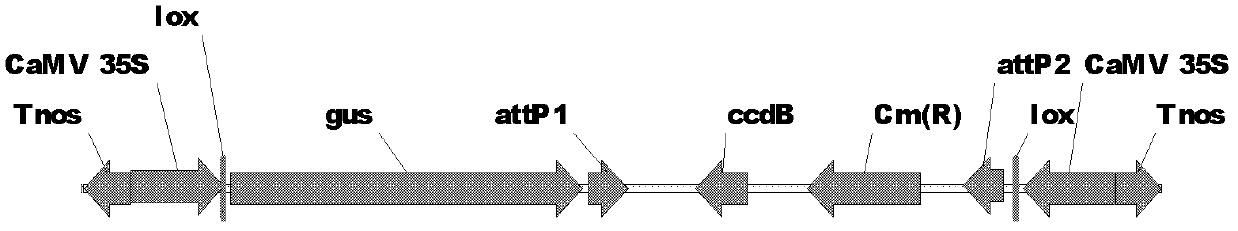
[0017] Example 1 Construction of Conjugated Promoter Plant RNAi Binary Expression Vector Containing Lox and attP Sites
[0018] 1. Mutation of Xho I restriction site of pTF101.1 vector
[0019] (1) Design the amplification primers as follows:
[0020] pTF 101.1FP: 5'-CTGGTCGACGTCCTTCTCAAATGAAATG-3'
[0021] pTF 101.1RP: 5'-GTCAATTGGAGCTCGGTACCCGGGGATC-3'
[0022] (2) Use the above primers to amplify the plasmid pTF101.1 as a template
[0023] The amplification system is as follows:
[0024] Pre-denaturation at 95°C for 3min, denaturation at 95°C for 30S, annealing at 52°C for 30S, extension at 72°C for 1min, 30 cycles, and finally extension at 72°C for 10min, and incubation at 4°C.
[0025] After recovering the target band amplified by PCR, it was digested with Sal I and Hind III to recover a 767bp fragment, which was connected between the Xho I and Hind III sites of the digested pTF101.1 vector to obtain a recombinant vector. Heat-shock transformed Escherichia coli DH5α ...
Embodiment 2
[0046] Example 2 Utilizing GCGS-1 Vector to Silence the Expression of GUS Gene in Transgenic Arabidopsis
[0047] 1. Obtaining transgenic Arabidopsis
[0048] (1) Add 0.1 μg of the constructed GCGS-1 carrier to 100 μL of Agrobacterium GV3301, transform by electric shock, add 900 μL of LB liquid medium, culture on a shaker at 28°C for 2 hours, centrifuge, remove 400 μL of the supernatant, and resuspend the bacteria The body was spread on LB solid medium containing 100mg / L Spe (specticomycin), 30mg / L Gen (gentamycin sulfate) and 100mg / L Rif (rifampicin), cultured at 28°C for 3 days, and screened Positive clone.
[0049] (2) Pick a single clone in 5mL liquid LB medium containing 100mg / L Spe, 30mg / LGen and 100mg / L Rif, culture it on a shaker at 28°C for 2 days, and inoculate it into LB liquid medium (containing 100mg / L Spe, 30mg / L Gen and 100mg / L Rif), shake culture at 28℃ until OD 600 When =0.5-0.8, the cells were collected by centrifugation, and resuspended in transformation ...
Embodiment 3
[0068] Example 3 Arabidopsis functional genome research based on long double-stranded RNA interference library
[0069] 1. Acquisition of cDNA library for functional genomics research
[0070] (1) Linearization of maize root cDNA library induced by low nitrogen
[0071] Construction of maize root cDNA library: Total RNA was extracted from maize roots treated with nitrogen deficiency for 2 days after 7 days of normal culture, and the mRNA was purified using the Poly(A) mRNA isolation and purification kit, and the full-length cDNA library construction kit ( Invitrogen, USA) constructed an entry clone library, and then used LR recombination to obtain a maize root cDNA library induced by low nitrogen.
[0072] Take 1 μL of the maize root cDNA library (1 μg / μL) induced by low nitrogen, add 1 μL of 10 × EcoRV buffer, 1 μL of EcoRV endonuclease, 6 μL of H 2 O, incubate at 37°C for 2h. Then add 40 μL H 2 O, 25 μL NaAc (3M), 187.5 μL absolute ethanol, store at -20°C overnight. Cen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com