Recombinant expression vector pEGFP (Plasmid Enhanced Green Florescence Protein)-DsRed-BP and application thereof to detection of activity of phiC31 integrase in mammalian cell
A technology of pegfp-dsred-bp and expression vector, which is applied in the fields of bioengineering and protein activity detection, can solve problems such as high detection cost and impact on transfection efficiency, and achieve high signal-to-noise ratio, simple and convenient implementation, and high fluorescence intensity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
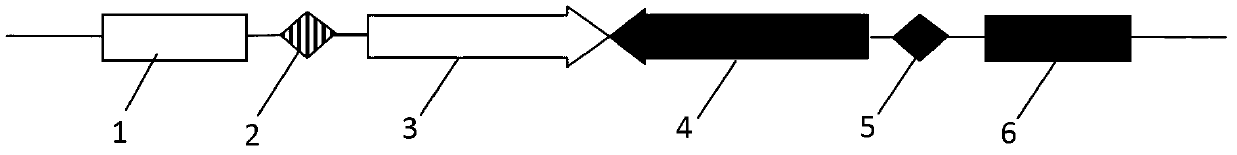
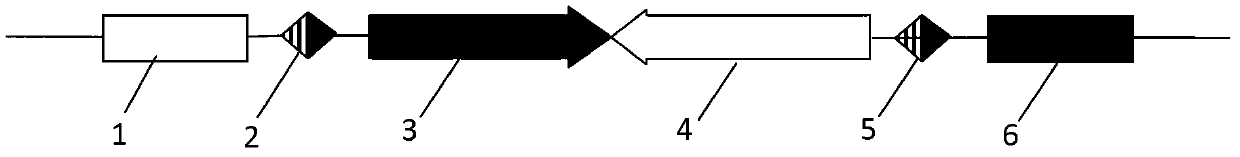
[0049] Embodiment 1: Construction of pEGFP-DsRed-BP vector
[0050] According to the minimum attB and attP sequences published in the literature, artificially synthesize the sequence containing the minimum attB and the reverse complementary sequence of attP, attB: CGGTGCGGGTGCCAGGGCGTGCCCTTGGGCTC CCCGGGCGCGTACTCCACC, attP reverse complementary sequence: GTAGTGCCCCAACTGGGG TAACCTTTGAGTTCTCTCAGTTGGGGGCGTAG, add Nhe Ⅰ at the left and right ends of the attB sequence and Age Ⅰ enzyme cutting sites, add BamH Ⅰ and Pst Ⅰ enzyme cutting sites at the left and right ends of the reverse complementary sequence of attP respectively. Using the pDsRed-monomer-N1 vector as a template, design the upstream primer Pred1 (EcoR Ⅰ): GGAATTCCACCGGTCGCCACCAT; the downstream primer Pred2 (Xho Ⅰ): CCGCTCGAGCGTAGAGTCGCGGCCGCTCT. Using the pair of primers synthesized above, carry out PCR amplification with the pDsRed-monomer-N1 vector as a template, and sequence and identify the amplified DsRed-monomer...
Embodiment 2
[0051] Example 2: Co-transfection of CHO, HEK293 and BHK cells with pCMVInt and pEGFP-DsRed-BP
[0052] CHO, HEK293 and BHK cells were seeded in six-well cell culture plates, and when the cell confluence reached 50%, 3 μg of plasmid (1.5 μg pCMVInt and 1.5 μg pEGFP-DsRed-BP) and 6 μl of Transfection reagent, mix and let stand for 30min. Aspirate the culture medium in the 6-well plate completely, wash it twice with serum-free culture medium, and wash away the serum. Add 1.5ml of serum-free culture medium to the mixture of plasmid and liposome, mix gently, carefully drop into a 6-well plate, and store at 37°C in 50ml / L CO 2 Incubate for 6 h in the incubator. Discard the transfection solution, add 2ml (DMEM containing 100ml / L serum) cell culture medium to continue culturing.
Embodiment 3
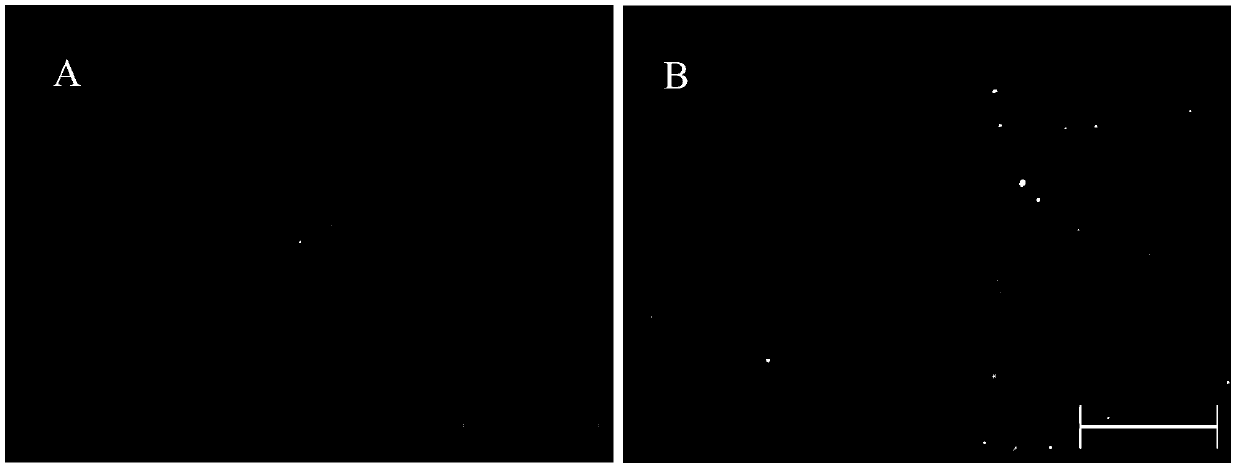
[0053] Example 3: Detection of fluorescent cells
[0054] After 24 hours of transfection, the above-mentioned cell culture plate was observed under a fluorescent microscope. It was found that DsRed was expressed in CHO, HEK293 and BHK cells, see the attached image 3 , 4 and 5. It indicated that φC31 integrase could play a role in these three kinds of cells. Fluorescent cells were digested, collected by centrifugation, and resuspended in PBS buffer for flow cytometry analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com