Esterolytic enzyme, coding gene, carrier, engineering bacterium and application of coding gene
An ester hydrolase and gene technology, applied in the field of ester hydrolase and its encoding gene, can solve the problems of complexity, difficulty in adoption, high cost and the like, and achieve the effects of high catalytic activity and stereoselectivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1: Cloning of the BMEST gene from Bacillus megaterium
[0029] The degenerate primer 1 and primer 2 were designed according to the DNA sequence of Bacillus megaterium DSM319 lipase (GenBank: CP001982.1).
[0030] Primer 1 sequence: ATGAATATCACAACGTTTGACG
[0031] Primer 2 sequence: TTATTTTTCTGTTGATATGCTTTTC
[0032] The genomic DNA of Bacillus megaterium (Bacillus megaterium) DSM319 was used as a template for PCR amplification (using TakaRa kit). The PCR reaction system and reaction conditions are shown in Table 1 and Table 2. Repeat steps 2 to 4 25 times.
[0033] Table 1: PCR amplification reaction system
[0034]
[0035] Table 2: PCR amplification reaction conditions
[0036]
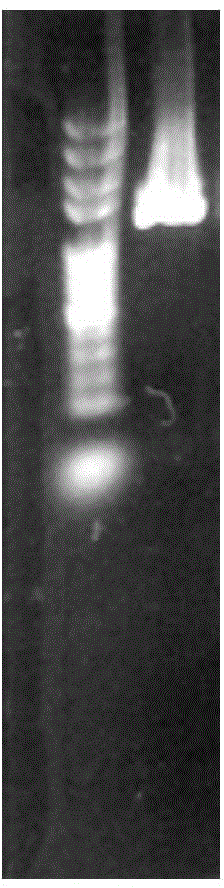
[0037] The PCR amplification product was detected by 0.8% agarose gel electrophoresis, and the product was a single band with a size of about 1400bp (such as figure 1shown). Purify and recover the PCR product. For specific steps, refer to the instructions of the Beijing Bi...
Embodiment 2
[0038] Embodiment 2: Construction of expression vector and transformant
[0039] A-T ligation of the target fragment with the pEASY-E1 expression vector, the ligation system is as follows in Table 3:
[0040] table 3: p EASY-E1Ex p Ression Vector connection system
[0041]
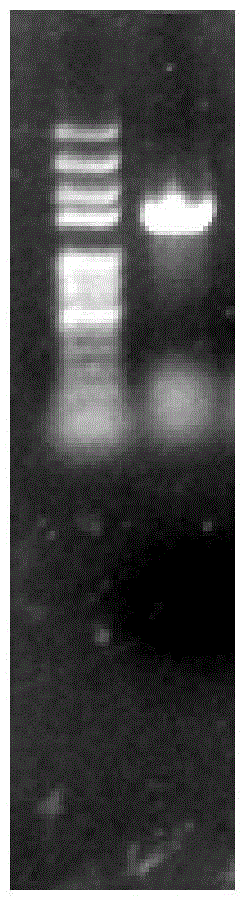
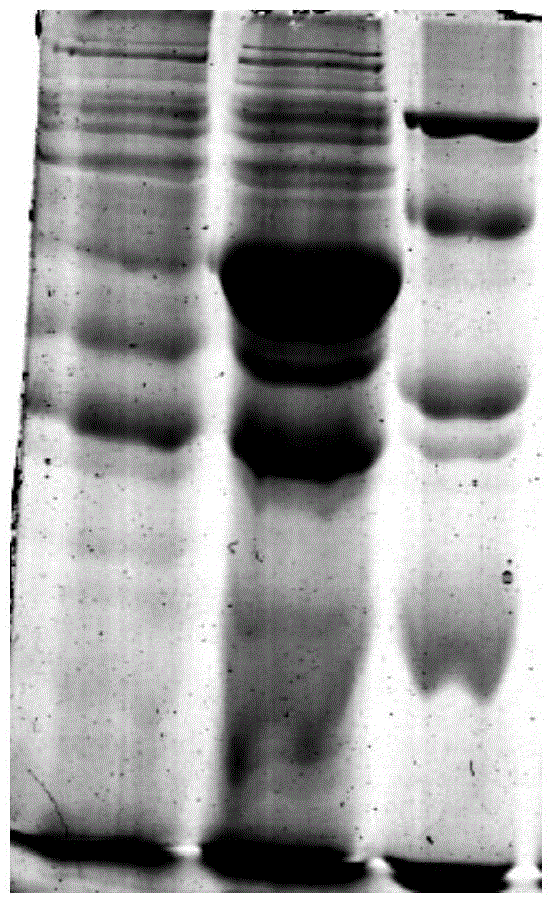
[0042] The connection system was reacted at 25°C for 15 minutes, and the connection product was added at 50 μ LClone in freshly thawed Trans-T1 competent cells, refer to pEASY-E1Expression Kit instructions for specific operations. PCR method to identify positive recombinants in the correct expression direction (such as figure 2 shown), transferred to LB medium to amplify the vector. The recombinant plasmid pEASY-E1-BMEST was extracted from Trans1-T1 cells with TaKaRa plasmid extraction kit, transformed into E.coli BL21(DE3) competent cells, and positive clones were obtained by screening with Amp-resistant medium and identified by PCR cloning. Contains the correct recombinant plasmid (such as im...
Embodiment 3
[0043] Example 3: Expression of Esterhydrolase
[0044] The genetically engineered strain E.coli BL21(DE3) / pEASY-E1-BMEST was inoculated in 50 mL of LB medium containing 80 μg / mL ampicillin, and cultured overnight at 37° C. with shaking. Take 500 μL of the culture solution and connect it to 50 mL of fresh LB medium containing 80 μg / mL ampicillin. When the OD600 is about 0.5-0.6, add IPTG until the concentration is 0.1 mmol / L to induce, and continue to culture at 25°C and 200 rpm for 10- After 12 hours, the target protein was overexpressed, and the cells were collected by centrifugation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com