PCR analyzing method for quantitatively detecting nucleic acid through RNA polymerase and ligase coupled reaction medium
A technology of RNA polymerase and analysis method, which is applied in the field of analysis and detection, can solve the problem of low sensitivity, and achieve the effect of good selectivity, simple operation process and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
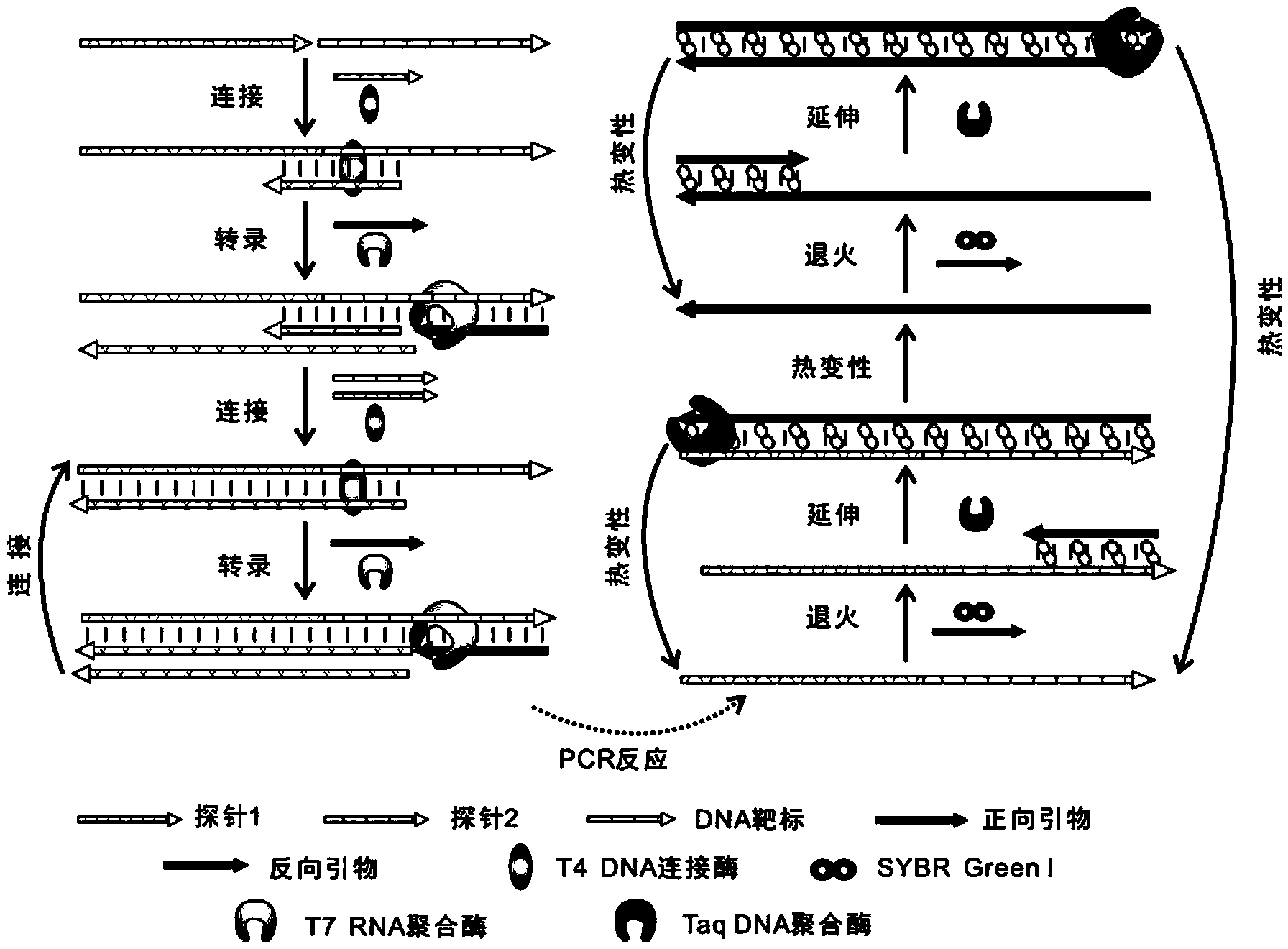
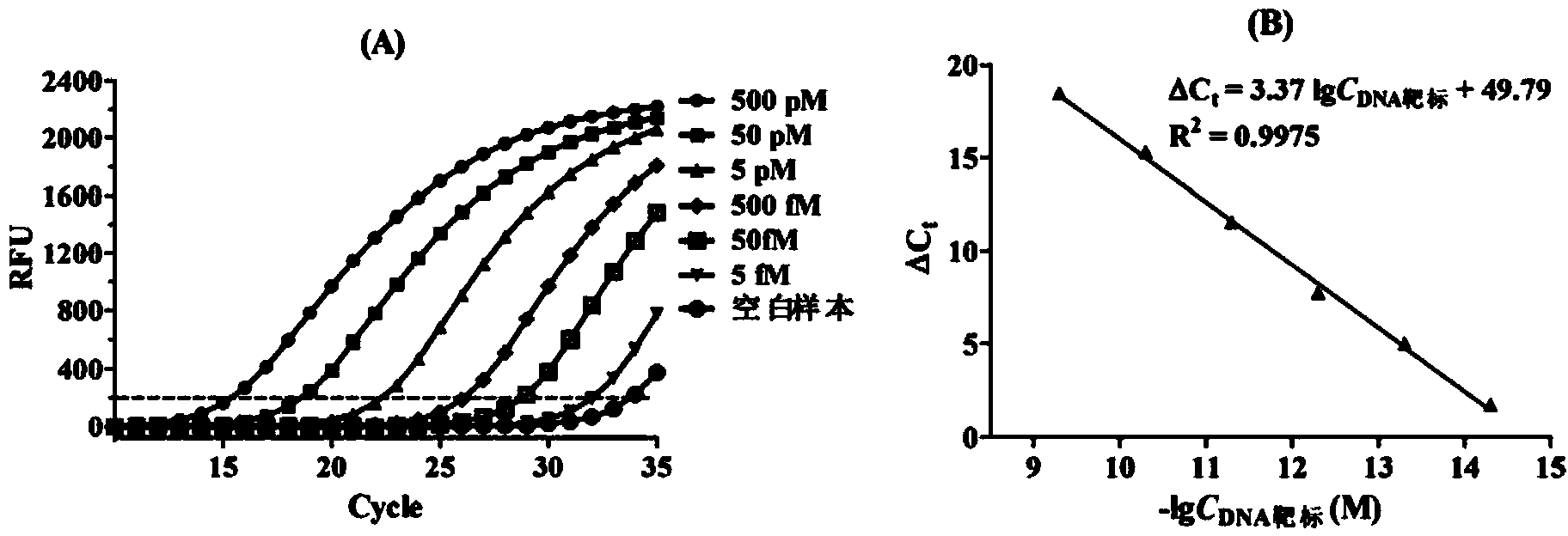
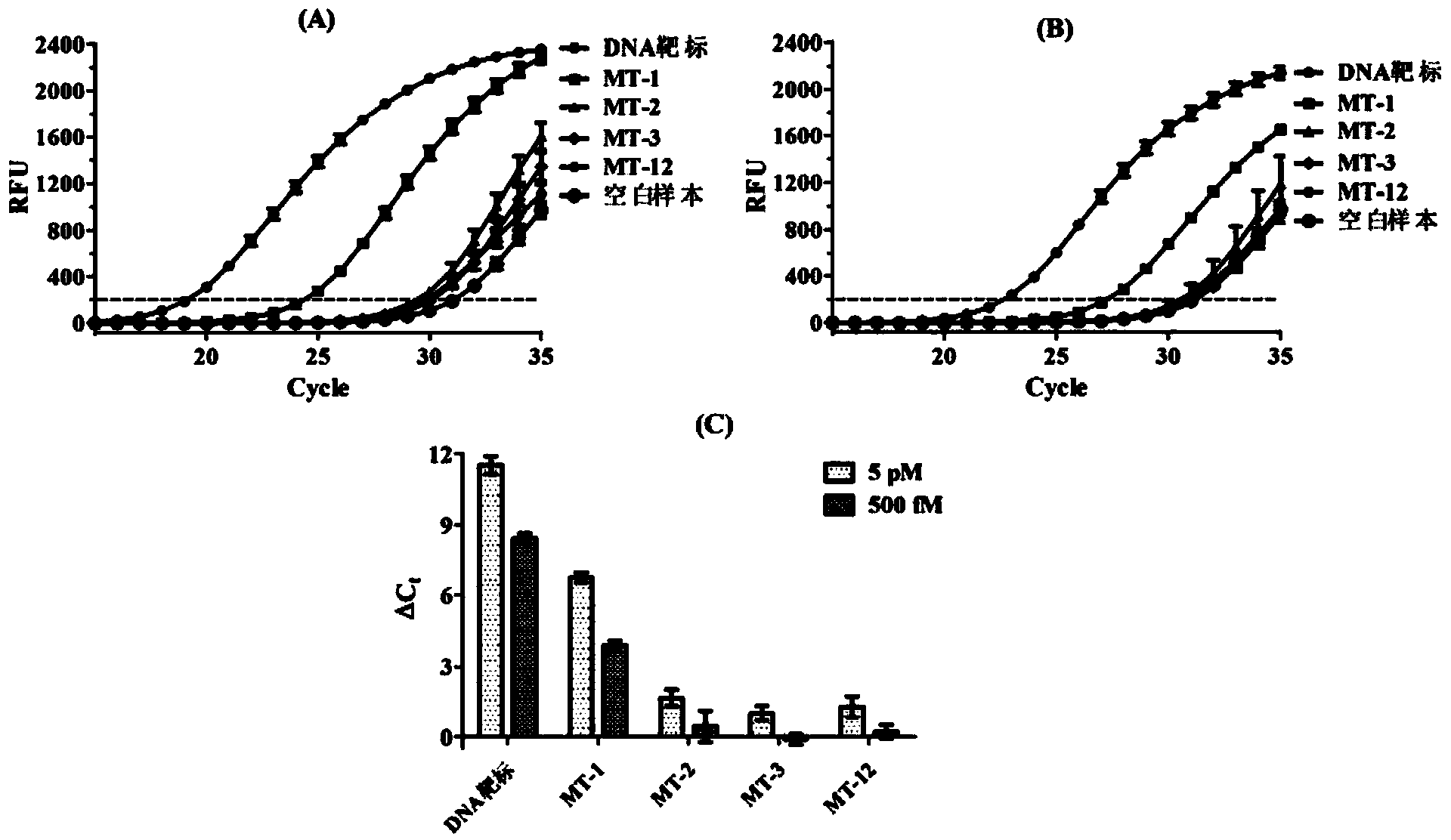
[0047] The analytical method for detecting the DNA target (5′-TTGAGGTGCATGTTTGTGCC-3′) by PCR mediated by T7 RNA polymerase and T4 DNA ligase coupling reaction, the detection principle is as follows figure 1 shown. The sequence complementary to the DNA target in probe 1 (5′-GGCACAAACAT-3′), the sequence complementary to the DNA target in probe 2 (5′-GCACCTCAA-3′) and the T7 promoter sequence (5′-CCCTATAGTGAGTCGTATTAGTGATC- 3').
[0048] The following specific implementation of DNA target detection will further illustrate the present invention. For the experimental methods that do not indicate the specific conditions, usually follow the conventional conditions or the conditions suggested by the manufacturer. The specific operation steps are as follows:
[0049] First prepare the Part A ligation reaction solution. Add 1 μL of transcription buffer solution (20 mM Tris-HCl, 3 mM MgCl 2 , 5mM DTT, 5mM NaCl and 1mM spermidine, pH 7.9), 1μL probe 1 solution (5nM), 1μL probe 2 so...
Embodiment 2
[0057] The analytical method for detecting miR-122 (5′-UGGAGUGUGACAAUGGUGUUUG-3′) by PCR mediated by T7 RNA polymerase and T4 RNA ligase 2 coupling reaction, the detection principle is as follows Figure 4 shown. Probe 1 sequence (5′-GGTATCCAGGGAAGTGGATACGAAGAATGCACAAACACCATTG-3′), probe 2 sequence (5′-(P)TCACACTCCACGCTGTGCCCTATAGTGAGTCGTATTAGTGATC-3′), forward primer sequence (5′-GGGAAGTGGATACGAAGAATGC-3′), reverse primer sequence ( 5'-GATCACTAATACGACTCACTATAGG-3').
[0058] The following specific implementation of target miR-122 detection will further illustrate the present invention. For the experimental methods that do not indicate the specific conditions, usually follow the conventional conditions or the conditions suggested by the manufacturer. The specific operation steps are as follows:
[0059] First prepare the Part A ligation reaction solution. Add 1 μL of transcription buffer solution (20 mM Tris-HCl, 3 mM MgCl 2, 5mM DTT, 5mM NaCl and 1mM spermidine, pH 7.9),...
Embodiment 3
[0066] The analytical method for detecting the DNA target (5'-TATTTGCCCCATTTTT-3') from respiratory syncytial virus (RSV) by PCR mediated by SP6 RNA polymerase and T4 DNA ligase coupling reaction, the detection principle is as follows Figure 5 shown. Probe 1 sequence (5′-GGTATCCAGGGAAGTGGATACGAAGCGTCGATAAAAATGG-3′), probe 2 sequence (5′-(P)GGCAAATACCTTTGCTTCGCTTCTATAGTGTCACCTAAAT-3′), forward primer sequence (5′-ATTTAGGTGACACTATAGAAGCG-3′), reverse primer sequence ( 5'-AGGGAAGTGGATACGAAGC-3').
[0067] The following specific implementation of DNA target detection will further illustrate the present invention. For the experimental methods that do not indicate the specific conditions, usually follow the conventional conditions or the conditions suggested by the manufacturer. The specific operation steps are as follows:
[0068] First prepare the Part A ligation reaction solution. Add 1 μL of transcription buffer solution (20 mM Tris-HCl, 3 mM MgCl 2 , 5mM DTT, 5mM NaCl and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com