Multichannel array type DNA (Deoxyribose Nucleic Acid) sequencing system and sequencing method thereof
A DNA sequencing and array technology, applied in the field of nanopore DNA sequencing system, can solve the problems of increasing solution viscosity, long time, lowering driving voltage, etc., to achieve the effect of reducing time cost, improving signal-to-noise ratio, and improving signal-to-noise ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0023] In order to make the object, technical solution and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with specific embodiments and with reference to the accompanying drawings.
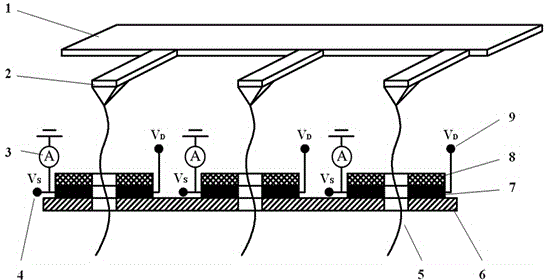
[0024] Such as figure 1As shown, the multi-channel array DNA sequencing technology integrating atomic force microscope and molybdenum disulfide field effect tube provided by the present invention consists of a current detection unit, a molybdenum disulfide field effect tube, an atomic force microscope feeding system and an array unit. The current detection unit is composed of an ammeter (3); the molybdenum disulfide field effect transistor is composed of a source power supply ( V s )(4), the drain supply ( V D ) (9), a silicon nitride substrate (6), a single-layer molybdenum disulfide nanoribbon (7) with nanopores and an insulating layer (8). The AFM feeding system consists of the AFM probe base (1) and the AFM probe ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com