Halohydrin dehalogenase mutant from parvibaculum lavamentivorans and application of halohydrin dehalogenase mutant
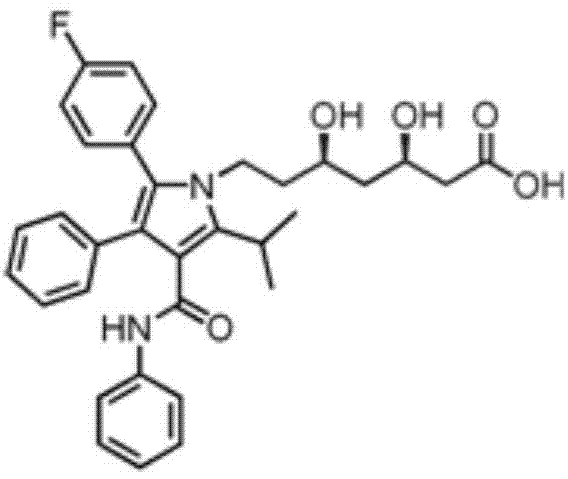
A technology of halohydrin dehalogenase and mutants, which is applied in the field of preparation of atorvastatin intermediate-4-cyano-3-hydroxybutyrate ethyl ester, can solve the problem of heavy drug burden for patients, high price, and Problems such as staying high
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: Construction of random mutation library of halohydrin dehalogenase
[0039] Random mutation technology reference (Current Protocols in Protein Science 26.6.1-26.6.10, 2011; Anal. Biochem. 2008, 375: 376-378) description. Primer P1 and primer P2 were designed according to GenBank ABS64560.1 gene sequence (see Table 1). First, an error-prone PCR experiment is performed on the parental HHDH-PL gene (the nucleotide sequence is shown in SEQ ID NO.1) with primers P1 and P2. 50μL error-prone PCR reaction system: 25μL 2×GoTaq Master Mix, 5 μL 50 mM MgCl 2 buffer, 5μL 1mM MnCl 2 , 0.5 μL P1 (50 μM) and P2 (50 μM), 1 μL (100-200 ng) HHDH-PL plasmid template, 13 μL deionized water. PCR program: pre-denaturation at 95°C for 3 minutes, 25 cycles: 94°C for 30s, 55°C for 30s, 72°C for 60s, and finally extension at 72°C for 10 minutes. After the PCR products were verified by agarose nucleic acid electrophoresis, the products were purified using PCR cleanup Kit. MEGAWH...
Embodiment 2
[0040] Example 2: Establishment of a high-throughput method for screening halohydrin dehalogenase mutants
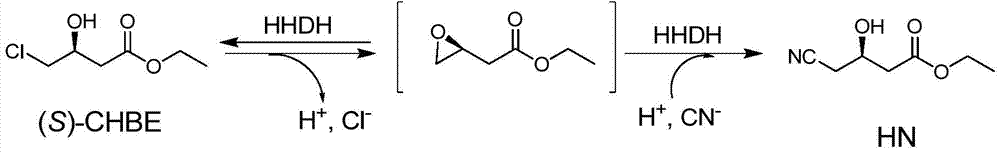
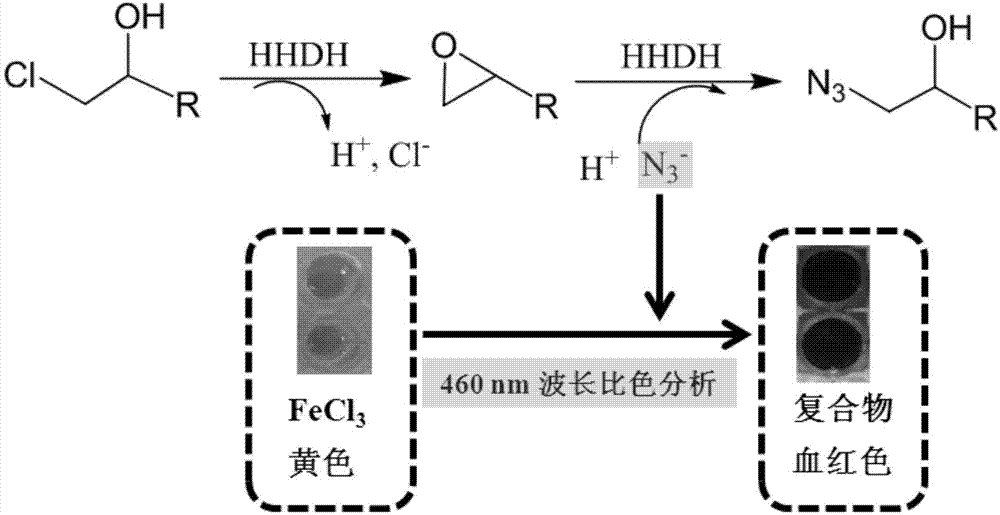
[0041] Since the purpose of this experiment is to screen a halohydrin dehalogenase mutant that can efficiently catalyze (S)-CHBE to synthesize HN, establishing an efficient high-throughput screening method is a key step to improve the realization of this invention. The methods reported in the literature to detect the activity of halohydrin dehalogenases are based on protons and halide ions produced by the dehalogenation process, but cannot detect the ring-opening process, so these methods are not suitable for screening halohydrin dehalogenation of (S)-CHBE to HN enzyme. In order to establish a suitable screening method, the present invention has established a new high-throughput screening method based on azide detection, such as image 3 shown.
[0042] Pick a single colony clone (the mutant library constructed in Example 1) and culture it in a 2 mL deep 96-well plate ...
Embodiment 3
[0043] Example 3: Construction of double mutants F176M / A187R and F176M / A187S
[0044] The plasmid of mutant F176M was extracted, and mutation primers P3 and P4 (see Table 1) were designed for A187R mutation, and mutation primers P5 and P6 (see Table 1) were designed for A187S mutation. Mutation system 50μL: 10μL 5×PS buffer, 4μL dNTPMix (0.8mM), 1μL (100-200ng) F176M plasmid, 0.5μL PrimerStar, 1μL (50μM) mutation primer and 32.5μL deionized water. PCR program: 72°C for 10 min, 95°C for 5 min, 30 cycles: 98°C for 10 s, 55°C for 15 s, 72°C for 8 min and 72°C for 10 min. After PCR was positive by 0.9% agarose gel electrophoresis analysis, take 20 μL of PCR solution, add 1 μL DpnI, digest at 37°C for 2 hours to remove template plasmid DNA, inactivate at 65°C for 10 minutes, and transform into competent cells E.coli BL21(DE3) , Spread LB plates containing ampicillin (50 mg / L). A single colony was picked and sequenced to analyze that the mutants F176M / A187R and F176M / A187S were su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com