Gossypium raimondii functional centromere sequence and molecular marker of same
A technology of functional centromere and Raymond's cotton, applied in the fields of bioinformatics and molecular cytogenetics, can solve the problems of undetermined DNA sequence CENH3 binding, the authenticity of the functional centromere sequence is questionable, etc. Real and reliable, good primer specificity, single band effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
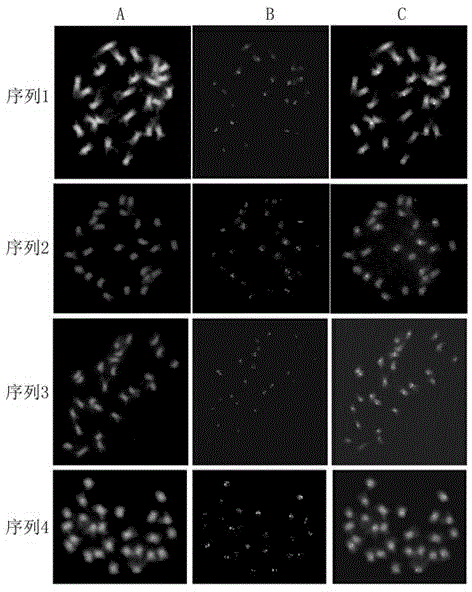
[0019] Example 1: Implementation of ChIP-Seq Technology
[0020] ChIP-Seq technology refers to the combination of chromatin immunoprecipitation ChIP and next-generation sequencing. This method is feasible and stable. It is not only suitable for the study of the binding relationship between a single gene and protein, but also for the genome-wide study of specific protein binding sites.
[0021] The basic process of ChIP-Seq used in the present invention is:
[0022] 1. Grind fresh and young leaves with liquid nitrogen; 2. Extract cell nuclei and enzymatically hydrolyze them with micrococcal ribozyme; 3. Add the antibody of centromere-specific histone CENH3 to combine with the target protein-DNA complex; 4. Add ProteinA binds to the antibody-target protein-DNA complex and precipitates it; 5. Washes the precipitated complex to remove some non-specific binding; 6. Elutes to obtain an enriched target protein-DNA complex; 7. 8. PCR analysis and preparation of sequencing library; 9....
Embodiment 2
[0039] Embodiment 2: Primer design and PCR amplification
[0040] Use the primer design software Primer Premier 5.0 to design primers for the functional centromere sequence. The set conditions are: first, the full-length or partial sequence of the nucleic acid sequence; second, the length of the primer is 21 bp±5 bp; third, the GC content of the primer sequence 40-60%; Fourth, the PCR product size is 200-700 bp.
[0041] The conditions of PCR amplification were: pre-denaturation at 95°C for 3 minutes, 35 cycles at 95°C for 30s, 47-53°C for 30s, 72°C for 45s, and a total extension at 72°C for 7 minutes. It should be noted that different primer sequences have different annealing temperatures: 50°C for sequence 1, 47°C for sequence 2, 53°C for sequence 3, and 51°C for sequence 4. The band amplified by PCR was single and bright. Therefore, the QIAGEN kit was directly used to purify and recover the PCR product, its product number: 28104, and its name: QIAquick PCR Purification Ki...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com