Method for enhancing glucose conversion rate of Aspergillus niger saccharifying enzyme
A technology of Aspergillus niger and conversion rate, which is applied in the field of genetic engineering to achieve the effect of reducing production costs and increasing the conversion rate of starch and sugar
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
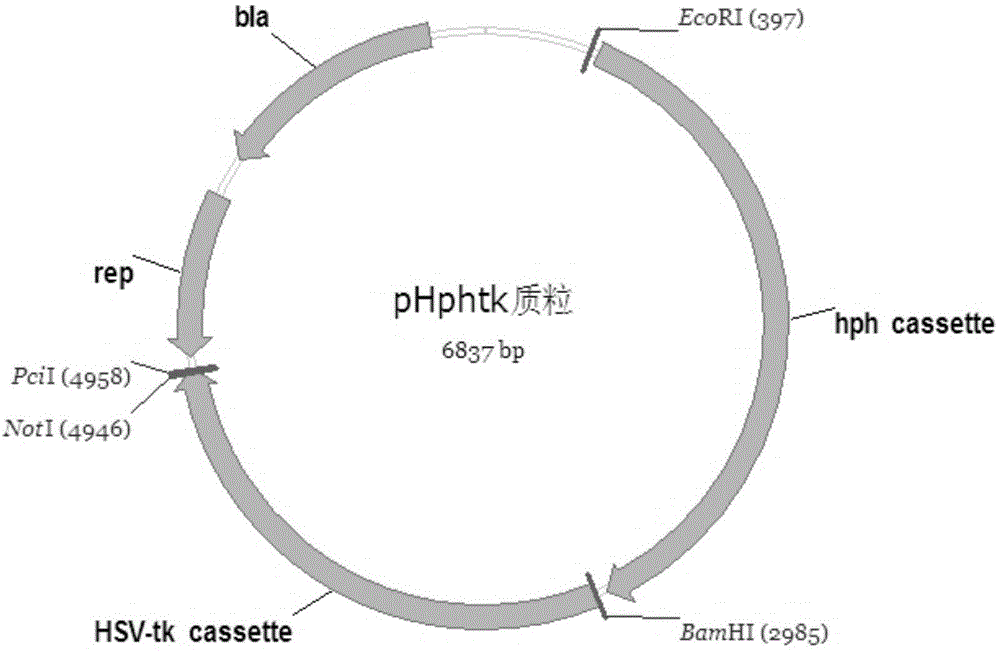
[0022] The construction of embodiment 1 pHphtk plasmid
[0023] The plasmid consists of the following 3 parts, constructed by GenScript company, see the plasmid map figure 1 .
[0024] (1) 2305bp fragment obtained after XbaI-PciI double digestion of pUC57 plasmid;
[0025] (2) hph gene expression cassette, the sequence is shown in SEQ ID NO.4;
[0026] (3) HSV-tk expression cassette, the sequence is shown in SEQ ID NO.5.
Embodiment 2
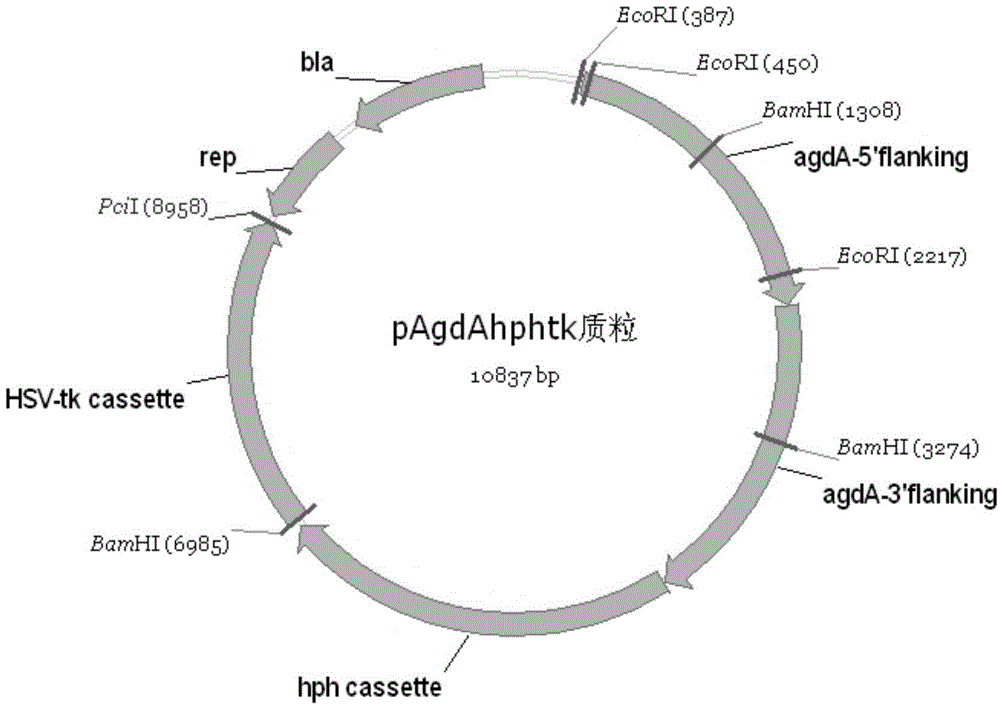
[0027]Example 2 Construction of integrated plasmids pAgdAhphtk and pAgdBhphtk
[0028] The integrated plasmid used for agdA gene knockout is called: pAgdAhphtk, and its construction method is as follows:
[0029] The pHphtk plasmid was linearized by vector-F and vector-R primers; at the same time, agdA-5'-F and agdA-5'-R, agdA-3'-F and agdA-3'-R were used as primers, respectively, to Aspergillus niger genomic DNA was used as a template, and a 2kb DNA fragment flanking the 5' end of agdA and a 2kb DNA fragment flanking the 3' end of agdA with a 40bp recombination arm were amplified by PCR; , 2kb DNA fragment flanking the 3' end of agdA by Gibson Master Mix Kit (E2611, New England Biolabs) was recombined to obtain the integrated plasmid pAgdAhphtk, see the plasmid map figure 2 .
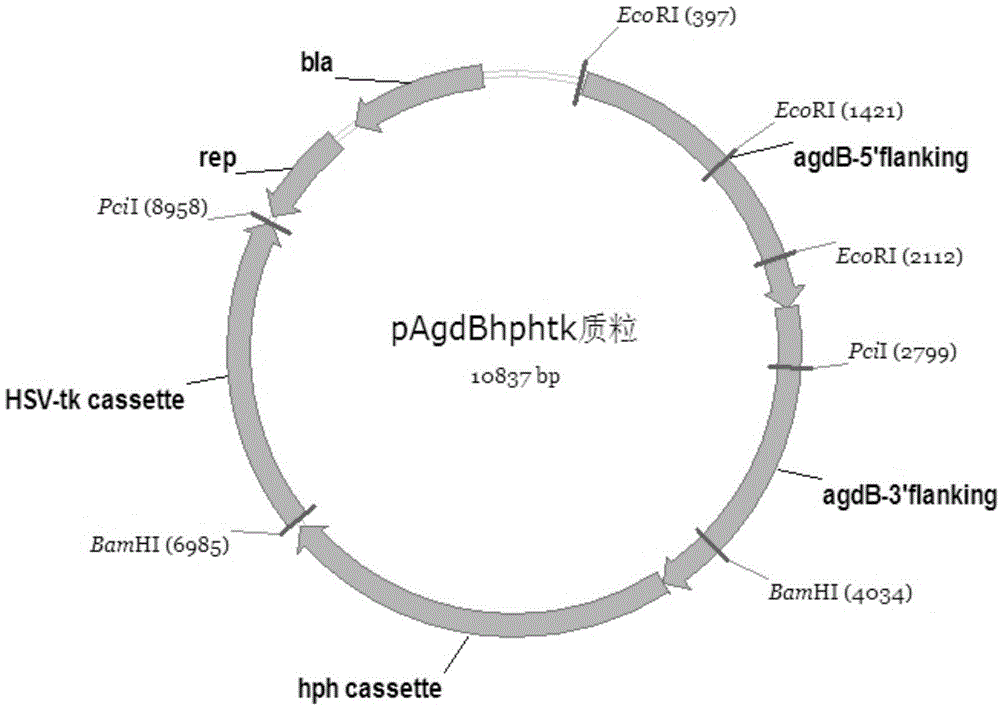
[0030] The integrated plasmid used for agdB gene knockout is called: pAgdBhphtk, and its construction method is as follows:
[0031] The pHphtk plasmid was linearized by vector-F and vector-R pri...
Embodiment 3
[0035] Embodiment 3 Transformation of pAgdAhphtk / pAgdBhphtk plasmid
[0036] The starting strain of this example is CBS513.88, which was purchased from the Netherlands Fungal Culture Collection (CBS-KNAW Fungal Biodiversity Centre).
[0037] Using the protoplast transformation method, the specific steps are as follows:
[0038] Preparation of protoplasts: Aspergillus niger mycelium was cultured in nutrient-rich TZ liquid medium (0.8% beef extract powder; 0.2% yeast extract; 0.5% peptone; 0.2% NaCl; 3% sucrose; pH5.8). The mycelium was filtered from the culture medium through mira-cloth (Calbiochem Company) and washed with 0.7M NaCl (pH 5.8). Sigma) and lysozyme (Sigma) 0.2% enzymatic hydrolysis solution (pH5.8), 30° C., 65 rpm for 3 h. Then place the enzymatic solution containing protoplasts on ice and filter with four layers of lens-cleaning paper. After the filtrate is centrifuged at 3000rpm, soft & 4°C for 10min, the supernatant is discarded; the protoplasts attached to t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com