Real-time fluorescence quantification PCR method with magnetic bead nucleic acid extraction and amplification conducted in one tube
A real-time fluorescence quantitative and nucleic acid extraction technology, applied in the field of molecular biology, can solve the problems of unstable nucleic acid cleavage efficiency, nucleic acid loss, nucleic acid pollution, etc., and achieve the effect of optimized nucleic acid extraction effect, low cost, and complete protein removal
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
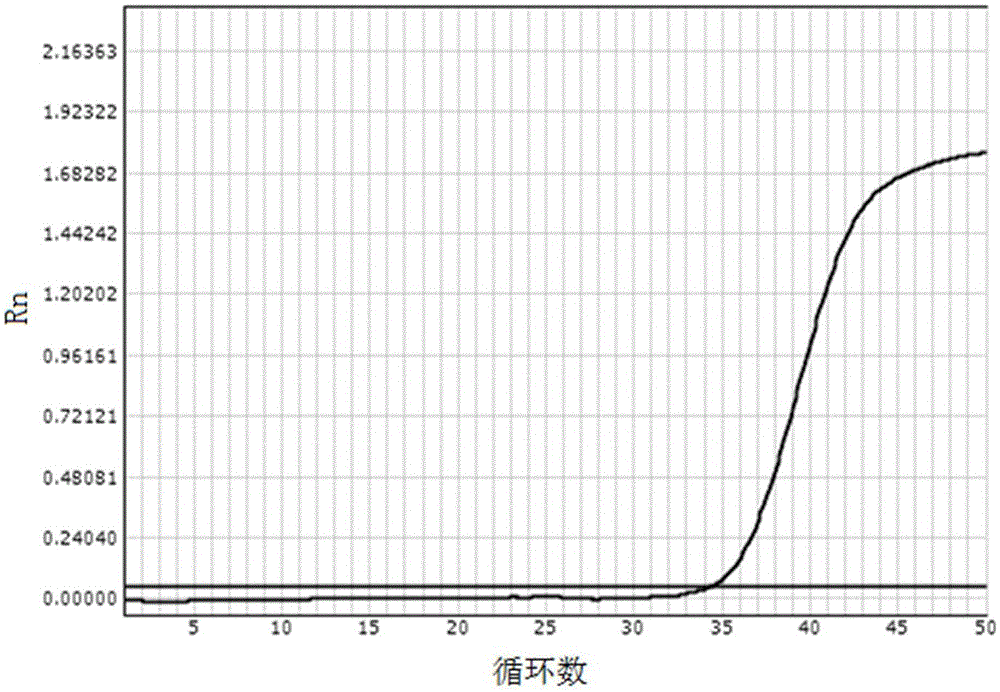
[0075] Embodiment 1: The real-time fluorescent quantitative PCR detection of HBVDNA
[0076] The reagents prepared according to the above-mentioned lysate scheme 1) and washing solution scheme 1) mixed with magnetic beads were used respectively for the clinical diagnosis and the tested HBVDNA quantitative value of 2×10 1 IU / ml hepatitis B patient's serum carries out the operation of following steps:
[0077] (1) Dispensing of PCR lysate: Dispense the prepared lysate mixed with magnetic beads into dedicated PCR tubes at a rate of 100 μl per tube.
[0078] (2) Adding samples: Take 100 μl of serum and add it to the above-mentioned PCR tube containing the lysate mixed with magnetic beads, gently blow and mix 5 times with a tip, and let stand at room temperature for 10 minutes.
[0079] (3) Aspirate and discard the liquid: Place the above PCR tubes on the eight-row magnetic stand, let it stand for 2 minutes, and use a pipette to absorb the liquid on the opposite side of the magnet...
Embodiment 2
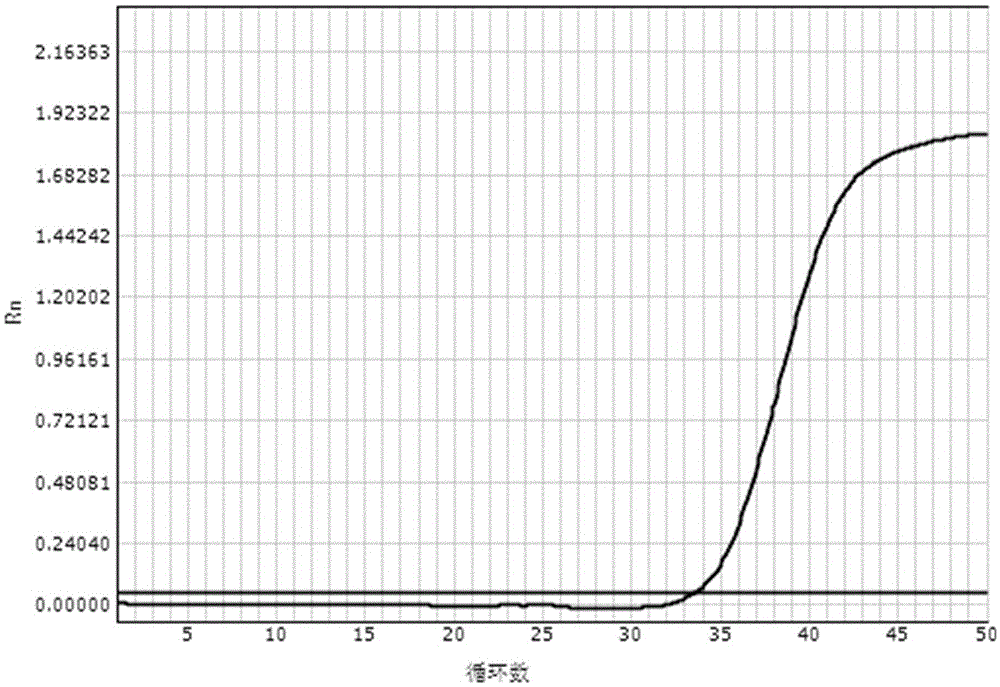
[0087] Embodiment 2: The real-time fluorescent quantitative PCR detection of HBVDNA
[0088] The reagents prepared according to the above-mentioned lysate scheme 2) and washing solution scheme 2) mixed with magnetic beads were used respectively for the clinical diagnosis and the tested HBVDNA quantitative value of 2×10 1 IU / ml hepatitis B patient's serum carries out the operation of following steps:
[0089] (1) Dispensing of PCR lysate: Dispense the prepared lysate mixed with magnetic beads into dedicated PCR tubes at a rate of 100 μl per tube.
[0090] (2) Adding samples: Take 100 μl of serum and add it to the above-mentioned PCR tube containing the lysate mixed with magnetic beads, gently blow and mix 5 times with a pipette tip, and let stand at room temperature for 5 minutes.
[0091] (3) Aspirate and discard the liquid: place the above-mentioned PCR tubes on the eight-row magnetic stand, let it stand for 2 minutes, and use a pipette to absorb the liquid on the opposite s...
Embodiment 3
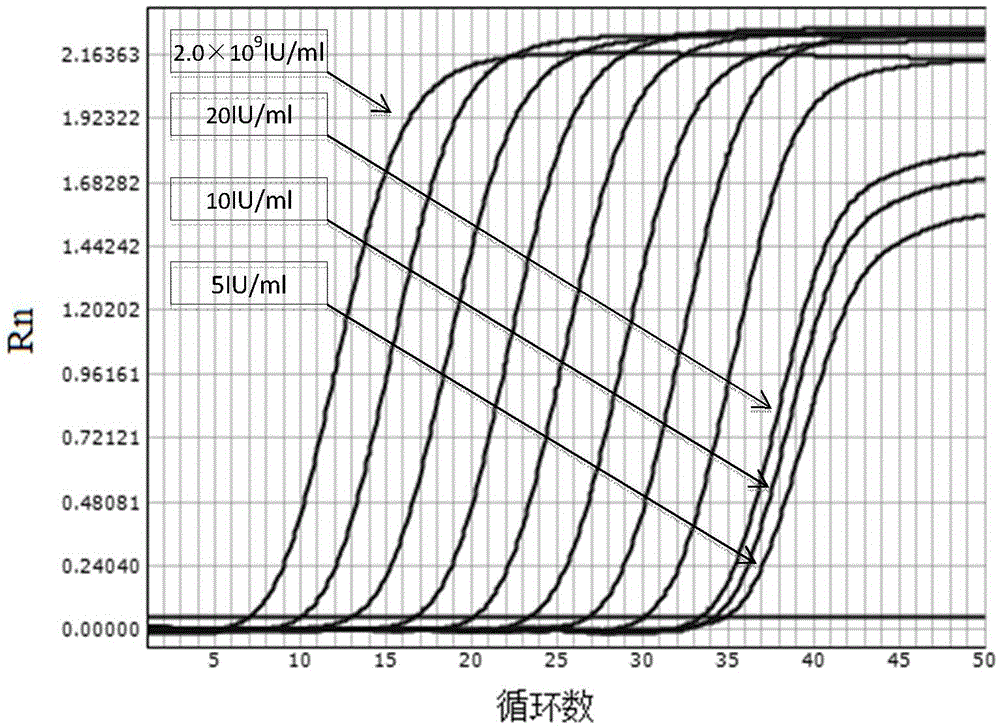
[0099] Embodiment 3: Sensitivity test of real-time fluorescent quantitative PCR method of the present invention
[0100] Serum samples negative for HBVDNA were used as diluent, and the clinical diagnosis was confirmed and the quantitative value of HBVDNA after testing was 2×10 9 The IU / ml serum of hepatitis B patients was diluted sequentially to obtain HBVDNA concentrations of 5IU / ml, 10IU / ml, 20IU / ml, and 2×10 2 IU / ml, 2×10 3 IU / ml, 2×10 4 IU / ml, 2×10 5 IU / ml, 2×10 6 IU / ml, 2×10 7 IU / ml, 2×10 8 IU / ml, 2×10 9 IU / ml of sample. The same method as in Example 1 was used to perform real-time fluorescent quantitative PCR detection on the above diluted samples.
[0101] Experimental results such as image 3 Shown, where the curve from right to left represents the concentration of 5IU / ml, 10IU / ml, 20IU / ml, 2×10 2 IU / ml, 2×10 3 IU / ml, 2×10 4 IU / ml, 2×10 5 IU / ml, 2×10 6 IU / ml, 2×10 7 IU / ml, 2×10 8 IU / ml, 2×10 9 IU / ml of sample. The results show that the present inventi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com