Identification and application of synthesis gene cluster of cordycepin
A cordycepin and gene cluster technology, applied in the field of genes, can solve the problem of unclear synthesis mechanism of cordycepin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0089] (1) Preparation of Cordyceps militaris conidia suspension: select fresh Cordyceps militaris cultured on a PDA (PotatoDextroseAgar, Difco) plate under light for a week, scrape an appropriate amount of colonies to 1ml sterile 0.05% Tween-20 aqueous solution, and vortex to isolate the bacteria Filament and conidia; remove mycelia by filtering with glass wool or three-layer lens-cleaning paper, and collect the filtrate; centrifuge the bacterial liquid at 12,000rpm for 1min to collect spores; discard the supernatant, and resuspend the spores with 1ml sterile water to remove residual nutrients ; 12,000rpm, 1min, centrifuge again to collect spores; repeat steps 4 to 5 once to completely remove residual nutrients on the surface of spores; Liquid concentration adjusted to 5×10 5 Spores / ml, used as a working solution for subsequent experimental operations. (2) Transformation of Cordyceps militaris: transfer the frozen AGL-1 bacteria that have been successfully transformed into t...
Embodiment 1
[0197] Example 1. Acquisition of potential gene clusters for cordycepin biosynthesis
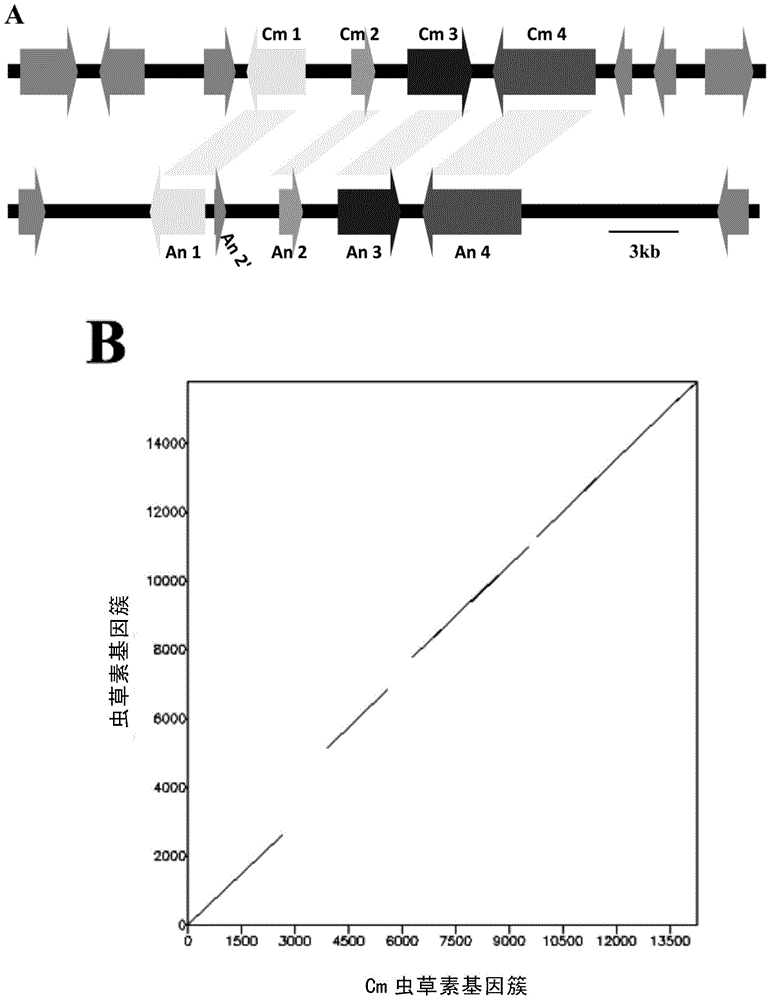
[0198] Through the comparative genome analysis of Cordyceps militaris (CM01 strain) and the model fungus Aspergillus nidulans (Aspergillus nidulans, A4 strain), the inventors have found the potential gene cluster (gene cluster) of cordycepin biosynthesis, which are respectively Cm1 (GenBank accession number : CCM_04436), Cm2 (CCM_04437), Cm3 (CCM_04438) and Cm4 (CCM_04439) ( figure 1 On A), the corresponding homologous genes in Aspergillus nidulans are An1 (AN3333), An2 (AN3331), An3 (AN3330) and An4 (AN3329) ( figure 1 A below).
[0199] Using the dotplot method to compare the sequences of the cordycepin synthesis gene clusters of these two species, it was found that the coding regions of each gene showed good collinearity ( figure 1 B). Structural and functional domain analysis showed that three genes in this gene cluster encode related metabolic enzymes, namely, Cm1 encodes oxidordeuct...
Embodiment 2
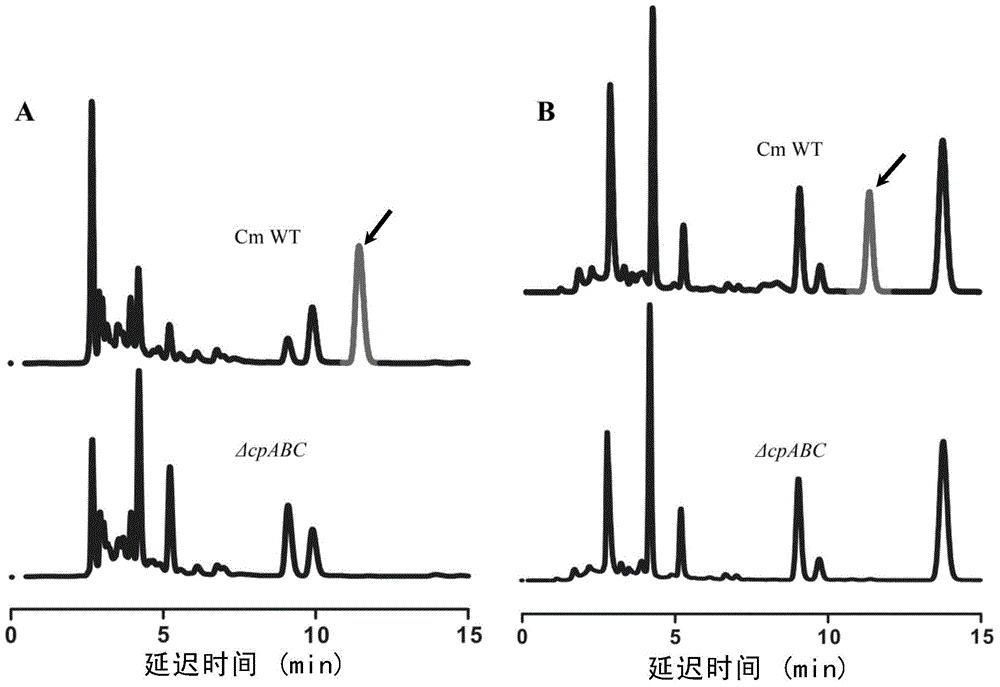
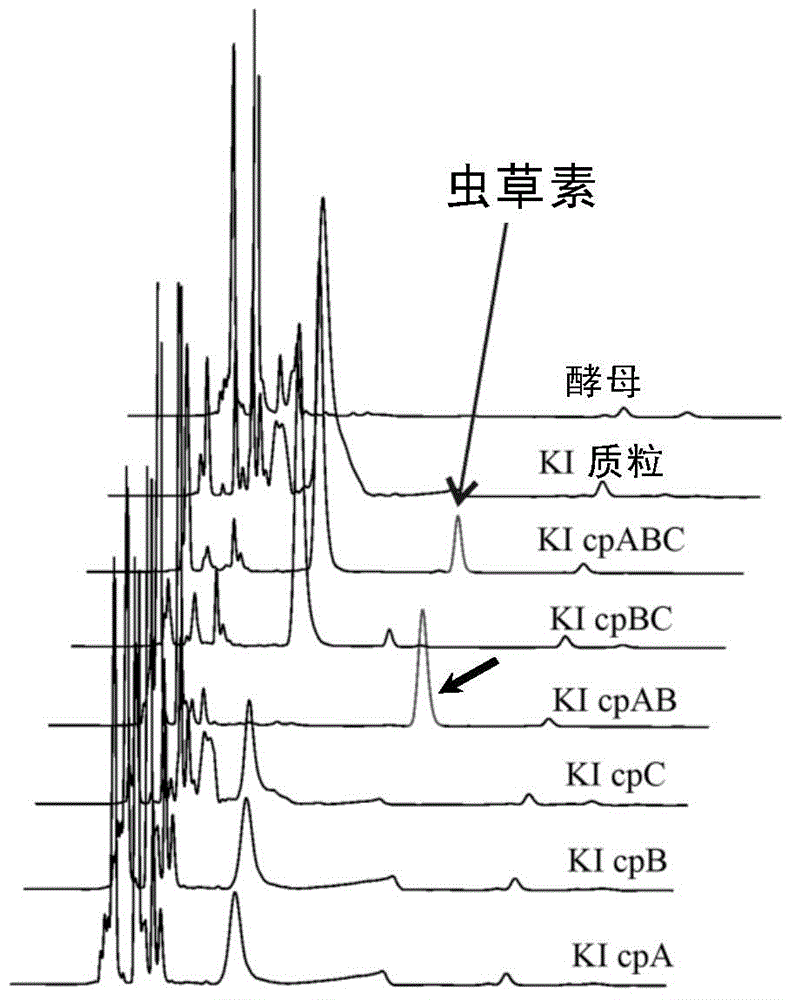
[0203] Embodiment 2, Cordyceps militaris wild strain CM01 and the comparison of cpABC knockout mutant producing cordycepin
[0204] For the potential gene clusters obtained from the above analysis, the inventors firstly knocked out three enzyme genes cpA, cpB and cpC in the genome of the wild strain CMO1 of Cordyceps militaris.
[0205] 1. Construction of gene knockout vector
[0206] The construction of Cordyceps militaris gene deletion mutants relies on the gene replacement method based on homologous recombination and the fungal transformation technology mediated by Agrobacterium tumefaciens AGL-1. With the binary vector pDHt-Bar (DuanZ, ChenY, HuangW, ShangY, ChenP, WangC. (2013) Linkageofautophagytofungaldevelopment, lipidstorageandvirulenceinMetarhiziumrobertsii.Autophagy9 (4): 538-549) (containing glufosinate resistance gene (Bar), for The upstream homology arm-Bar-downstream homology arm structure is formed, and the corresponding segment gene in the genome is knocked o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com