Method for extracting genomic DNA (deoxyribonucleic acid) from whole blood on basis of improved immunomagnetic bead method
A genome and magnetic bead method, applied in the biological field, can solve the problems of large amount of reagents, long test time, low qualification rate, etc., and achieve the effect of small amount of reagents, reduced test cost and short time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
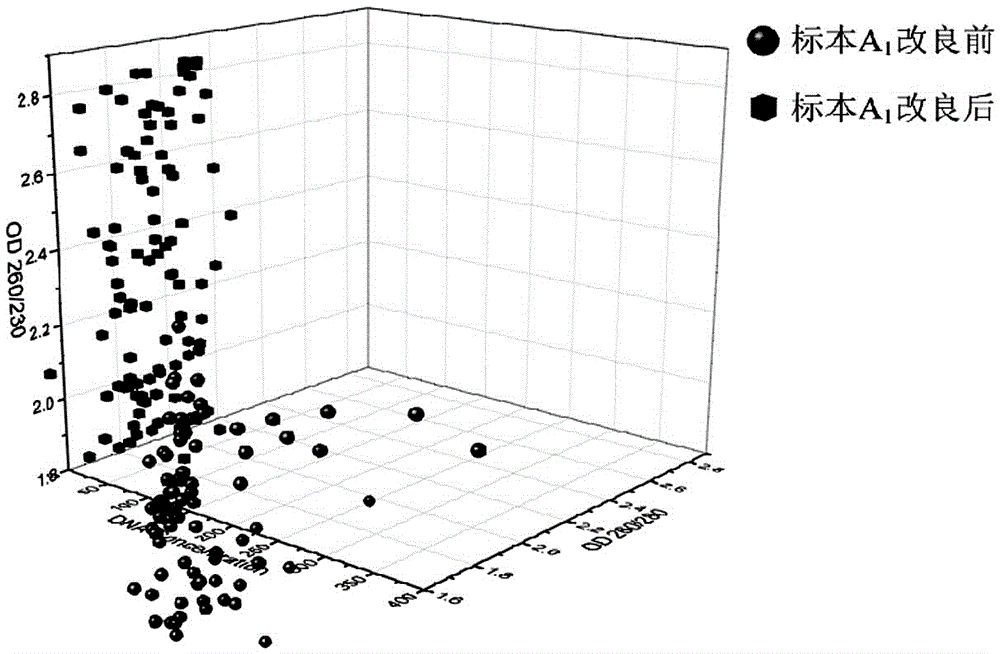
[0028] Whole blood sample A 1 Blood samples from stem cell donors in Hubei Branch of China Bone Marrow Bank.
[0029] Utilize the method of the present invention to above-mentioned whole blood sample specimen A 1 DNA detection, the specific steps are as follows:
[0030] (1) Red blood cell lysis
[0031] Add 500 μl of red blood cell lysate into a 2ml centrifuge tube, add 120 μl of completely thawed whole blood sample into the above centrifuge tube, oscillate fully, and mix until the cell fragments are evenly suspended and no clumps are seen. Dilute the lysate × 10,000 Rmp was centrifuged for 1 min, and the supernatant was removed.
[0032] (2) Leukocyte Lysis and DNA Adsorption
[0033] Dilute the proteinase K stock solution to a ratio of two times, draw 20 μl proteinase K and 140 μl LysisBuffer into the above centrifuge tube, and repeatedly pipette and mix 3 times, then incubate at 65°C, shake for 2 minutes and rest for 8 minutes, and alternately shake and stand for 3 cyc...
Embodiment 2
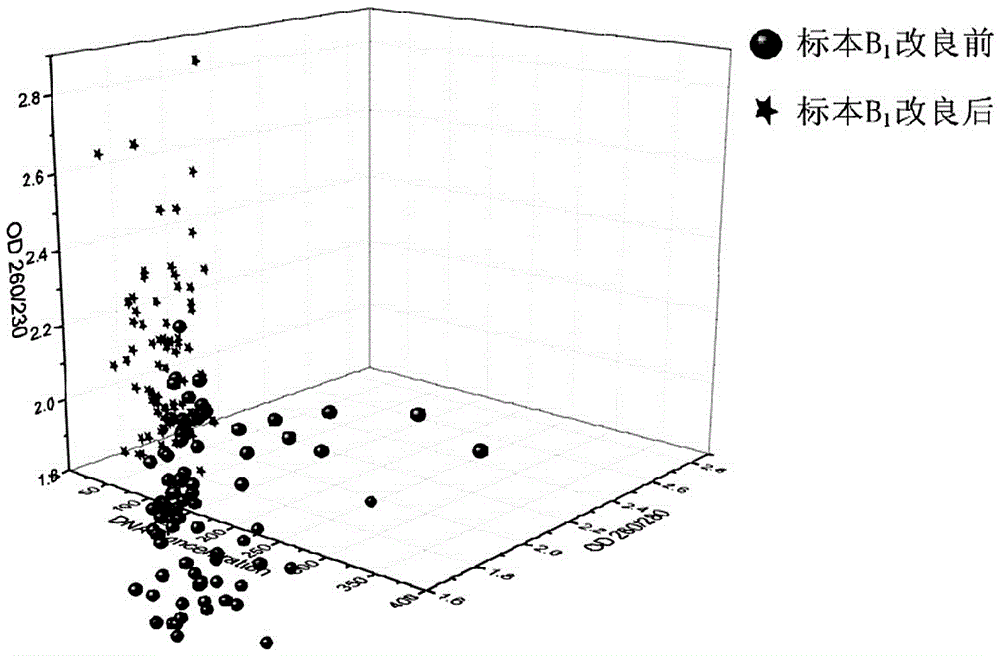
[0048] Whole blood sample B 1 Blood samples from stem cell donors in Hubei Branch of China Bone Marrow Bank.
[0049]Utilize the method of the present invention to above-mentioned whole blood sample specimen B 1 DNA detection, the specific steps are as follows:
[0050] (1) Red blood cell lysis
[0051] Add 480 μl of red blood cell lysate into a 2ml centrifuge tube, add 130 μl of completely thawed whole blood sample into the above centrifuge tube, oscillate fully, and mix until the cell fragments are evenly suspended and no clumps are seen. Dilute the lysate × 10,000 Rmp was centrifuged for 2min, and the supernatant was removed.
[0052] (2) Leukocyte Lysis and DNA Adsorption
[0053] Dilute the protease K stock solution in multiple ratios, pipette 20 μl proteinase K and 150 μl LysisBuffer into the above centrifuge tube, and repeatedly pipette and mix 3 times, then incubate at 65°C, shake for 2.5 minutes and rest for 6 minutes, and alternately shake and rest for 3 cycles; ...
Embodiment 3
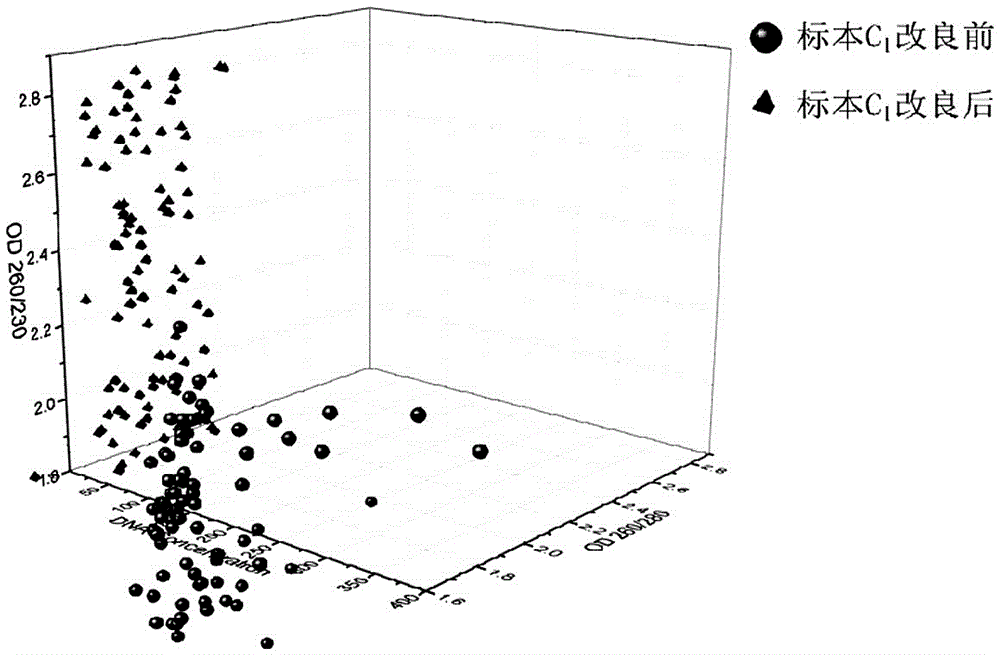
[0068] Whole blood sample C 1 Blood samples from stem cell donors in Hubei Branch of China Bone Marrow Bank.
[0069] Utilize the method of the present invention to above-mentioned whole blood sample C 1 DNA detection, the specific steps are as follows:
[0070] (1) Red blood cell lysis
[0071] Add 450 μl of red blood cell lysate into a 2ml centrifuge tube, add 150 μl of completely thawed whole blood sample into the above centrifuge tube, oscillate fully, and mix until the cell fragments are evenly suspended and no clumps are seen. Dilute the lysate × 10,000 Rmp was centrifuged for 3min, and the supernatant was removed.
[0072] (2) Leukocyte Lysis and DNA Adsorption
[0073] Dilute the proteinase K stock solution to a ratio of two times, draw 20 μl proteinase K and 120 μl LysisBuffer into the above centrifuge tube, and repeatedly pipette and mix 3 times, then incubate at 65°C, shake for 3 minutes and rest for 5 minutes, and alternately shake and stand for 3 cycles; add 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com