Hepatitis c virus NS3/4A protease, gene and method for detecting activity of inhibitor
A hepatitis C virus and protease inhibitor technology, which is applied in the field of anti-hepatitis C virus drug screening, can solve the problems of cumbersome and time-consuming cell models, difficult safety issues, difficult preparations, etc., and achieve high screening efficiency, low cost, avoid cumbersome effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
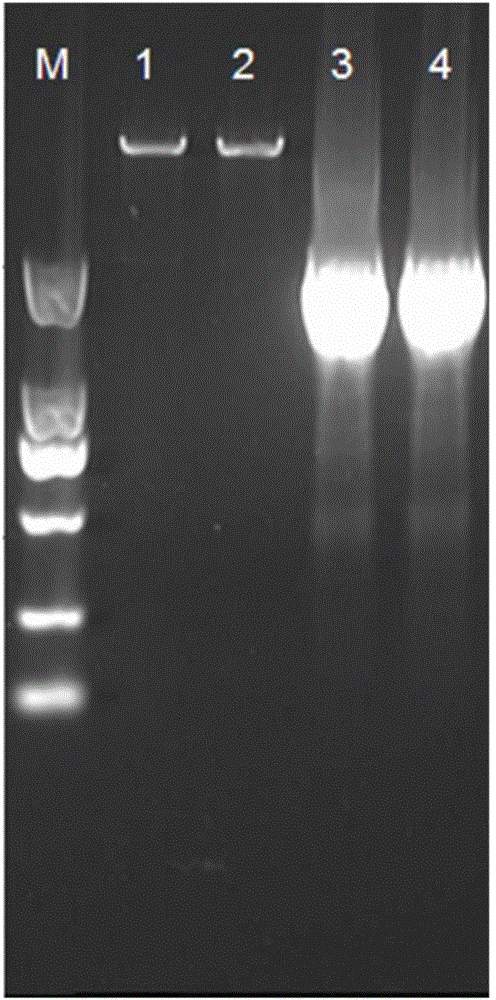
[0038] Construction of embodiment 1 recombinant plasmid pET28a-NS3
[0039] 1) A total of 164 complete gene sequences of all reported hepatitis C subtype 1b subtypes were obtained from the NCBI website, and cluster analysis was performed using http: / / www.drive5.com / uclust software, and the ones with a similarity of more than 90% were selected. The most representative sequence was used as the research object, namely the sequence HCV-1b / US / BID-V130 / 1994 (GenBank: EU155330.2). The entire sequence encoding NS3 / 4A is selected, the full length of the nucleic acid of the sequence is 2055bp, encoding 685 amino acids. In order to increase the in vitro activity and stability of the NS3 / 4A protease, the gene sequence was codon-optimized, and the nucleotide sequence of the optimized NS3 / 4A protease gene is shown in SEQ ID No.1 in the sequence table. The optimized gene was synthesized by chemical synthesis (synthesized by Shanghai Jierui Bioengineering Co., Ltd.), and the synthesized full...
Embodiment 2
[0044] Example 2 Construction, induced expression and protein purification of recombinant strains.
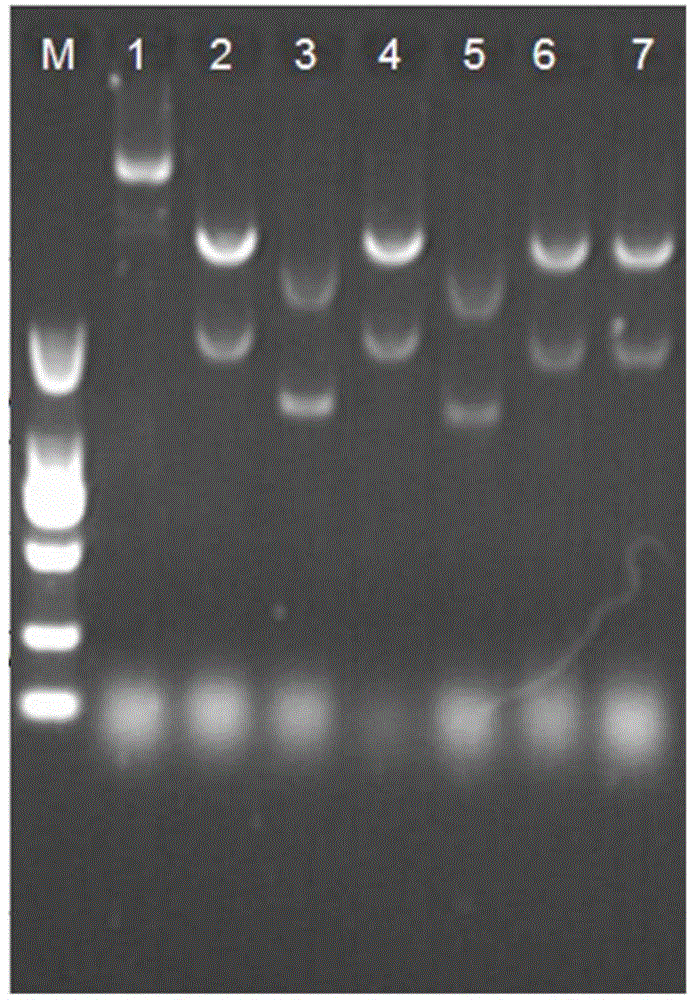
[0045] 1) The recombinant plasmid pET28a-NS3 was transformed into the expression host Escherichia coli BL21(DE3), spread on an LB agar plate containing 30 μg / mL kanamycin, and cultured at 37° C. for 12 hours. Pick a single colony grown on the resistant plate and inoculate it in 5 mL of LB liquid medium containing 30 μg / ml kanamycin, culture overnight at 200 rpm at 37°C, extract the plasmid, and carry out Bam HI and NdeI double enzyme digestion identification to obtain recombinant strains. Save the verified correct strain.
[0046] 2) Inoculate 50 μL of the correct single colony verified in step 1) into 5 mL of LB, and shake overnight at 200 rpm at 37° C. to obtain an overnight culture. Take 500μL overnight culture in 50mL LB, shake at 200rpm at 37℃ and culture to OD 600 When it is 0.6, add 0.5mM IPTG to induce, 18°C, 200rpm to induce culture for 22h, then collect the bacteri...
Embodiment 3
[0048] Activity experiment of embodiment 3NS3 / 4A protease
[0049] React 100 μg / mL NS3 / 4A protease with substrate P06221, add NS3 / 4A protease and substrate to the reaction system, and incubate. The reaction system is: NS3 / 4A protease solution 95 μL, 5 μM substrate 2.5 μL, incubated at 30°C for 60 minutes, set up 5 parallel controls, and adjusted the pH to 6.5, 7, 7.5, 8 and 8.5, respectively. Fluorescence-pH curves were measured under different pH conditions. The result shows that when measuring 60min, the fluorescence intensity is the highest when showing pH value 7.5 ( Figure 4 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com