Intracellular scaffold structure and method
A scaffold structure and inner scaffold technology, applied in the field of intracellular scaffold structure, can solve problems such as stability and scalability limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1 Construction method of TALE-DNA scaffold system prokaryotic expression plasmid
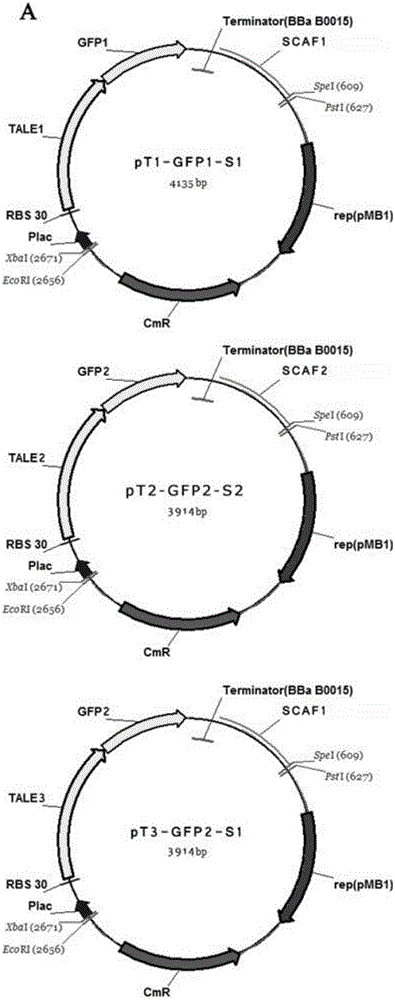
[0043] The TALE expression sequence was constructed using Golden Gate TALEN and TAL Effector Kit 2.0, and the DNA scaffold sequence was synthesized by DNA synthesis. Utilizing the properties of the restriction endonuclease Spe1 and Xba1 homologous enzymes, a multiple cloning site system of EcoR1, Xba1, Spe1 and Pst1 was designed on the pSB1C3 plasmid, and the Plac promoter, Plac promoter, and Nucleic acid elements such as ribosome binding site (RBS), TALE, split GFP, tryptophan monooxygenase (IAAM), indole acetamide hydrolase (IAAH), terminator (Ter) or DNA scaffold sequence (Scaffold) . Specifically construct the following plasmids: construct pSB1C3-Plac-RBS-TALE1-GFP1-Ter-Scaffold1 (pT1-GFP1-S1), pSB1C3-Plac-RBS-TALE2-GFP2-Ter-Scaffold2 (pT2-GFP2-S2) and pSB1C3- The Plac-RBS-TALE3-GFP2-Ter-Scaffold1 (pT3-GFP2-S1) plasmid was used to detect the binding ability of the TALE-GFP f...
Embodiment 2
[0044] Example 2 Verification of the binding ability of the TALE-GFP fusion protein provided by the present invention to the DNA scaffold
[0045] ChIP-PCR was used to test whether the fusion protein of TALE and heterologously expressed protein (GFP1 / 2) could effectively bind to the plasmid DNA scaffold. Firstly, the constructed pSB1C3-1, pSB1C3-2 and pSB1C3-3 plasmids were respectively transformed into Escherichia coli BL21 (DE3) strain competent, positive clones were screened, and inoculated in 5 mL of LB liquid medium containing ampicillin (50 μg / ml) , placed on a shaker at 37°C, and cultivated at 250rpm for 5 hours, then added the culture to LB liquid medium containing ampicillin (50 μg / ml) at a ratio of 1:100, placed on a shaker at 37°C, and continued to cultivate at 250rpm. Determination of bacterial growth curve, wait for OD 600 When the temperature reaches 0.6-1.0, add IPTG to make the final concentration 1mM, and then transfer to 20°C, 25°C or 30°C incubator at 200rp...
Embodiment 3
[0047] Example 3 Verification of the colocalization effect of the TALE-DNA scaffold system provided by the present invention on heterologous proteins
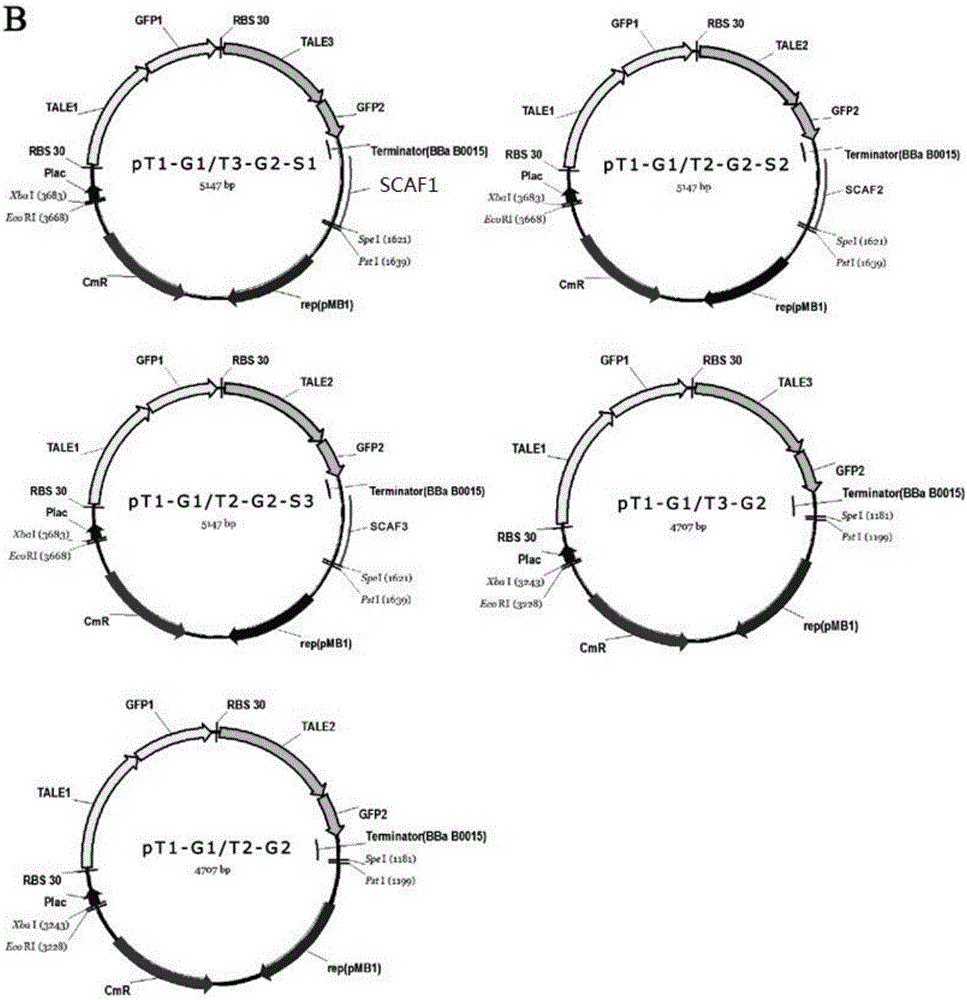
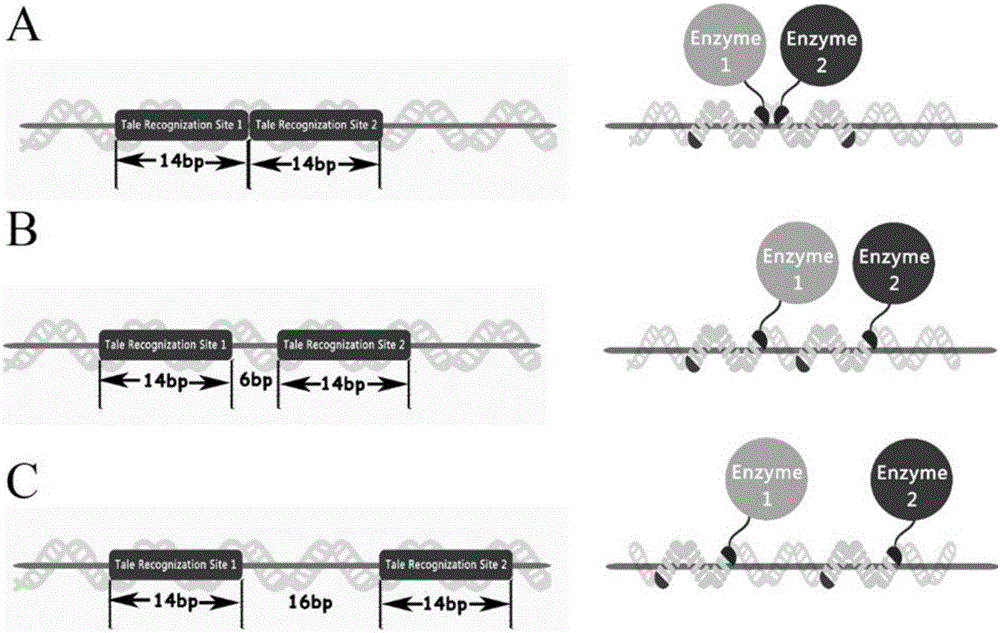
[0048] The colocalization effect of the TALE-DNA scaffold system provided by the present invention on heterologous proteins was verified by splitting GFP experiments. The five groups of plasmids pT1-G1 / T3-G2-S1, pT1-G1 / T2-G2-S2, pT1-G1 / T2-G2-S3, pT1-G1 / T3-G2, pT1-G1 / T2-G2 were respectively Transform Escherichia coli BL21 (DE3) strain to be competent, and after expanding the culture of positive clones, induce overnight at 25° C. with 1 mM IPTG. The group without IPTG was used as the uninduced control group. Bacterial samples were subsequently collected, and the OD of each group was measured with a fluorescent microplate reader Fluoroskan Ascent FL (Thermo Scientific Company) 600 and fluorescence intensity FI (excitation light wavelength: 488nm; emission light wavelength: 538nm). Such as Figure 4 As shown in B, FI / OD of pT1-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com