Detection kit and detection method for Cyprinid herpesvirus III
The technology of koi herpes virus and detection kit is applied in the detection field of fish pathogenic virus to achieve the effects of ensuring fish health, simple and easy operation procedures, and improved detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
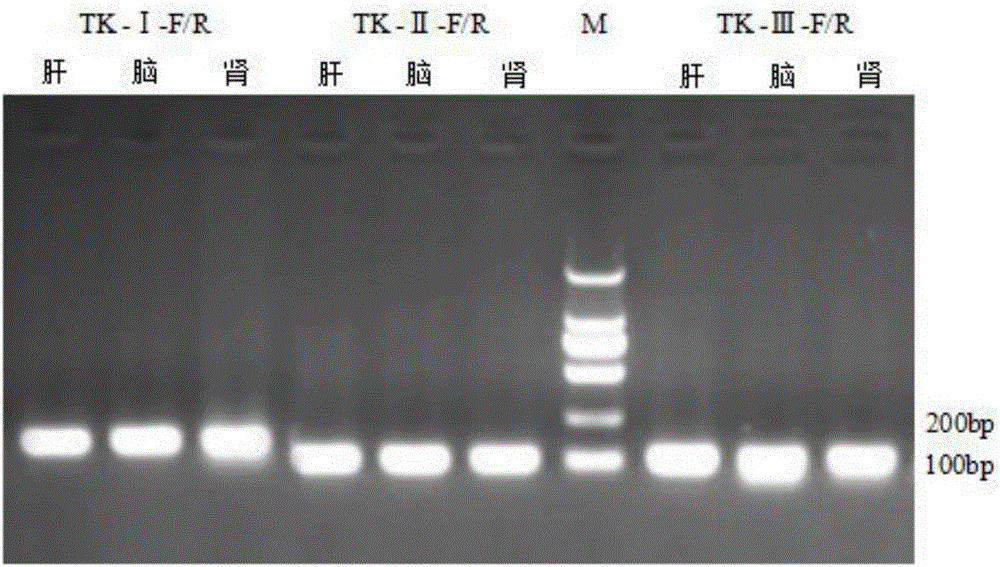
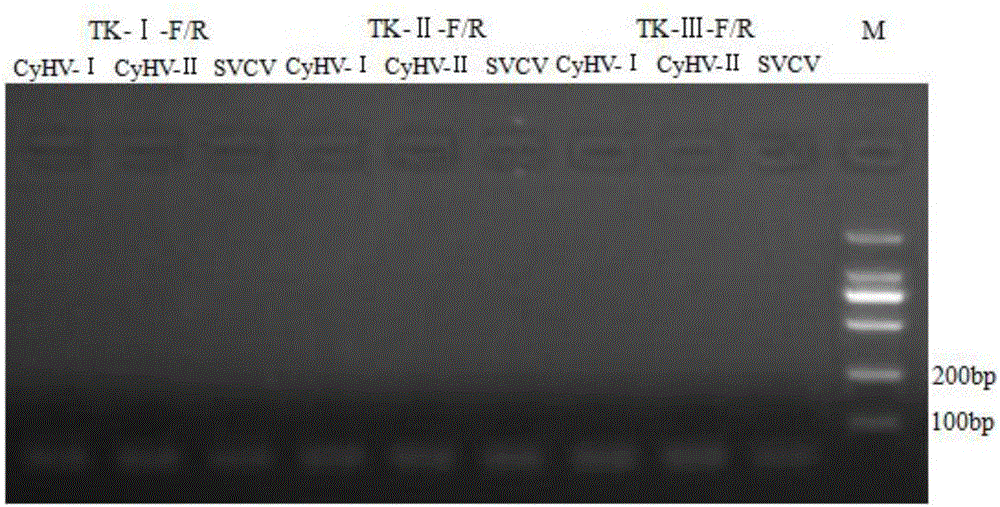
[0045] Example 1 Primer Design and Screening for Fluorescent Quantitative PCR Detection of Koi Herpesvirus Type III TK Gene
[0046] 1. Bioinformatics method to design primers and conduct primer screening
[0047] Download the full sequence of Koi Herpesvirus Type Ⅲ collected by GenBank (GeneBank: NC_009127.1), and also download the negative control virus genome, Koi Herpesvirus Type Ⅰ, Koi Herpesvirus Type Ⅱ, and Carp Spring Viremia Virus, please See Table 1. After the comparison by Clustal X, select the appropriate region to design primers. This region is specifically expressed in Koi herpesvirus type III, but there is at least 10%-30% difference compared to the negative control. After selecting the region, use ABI PrimerExpress 3.0 real-time fluorescence quantitative PCR primer design software, design synthetic primers. The extracted alternative primers were screened according to the following requirements:
[0048] 1) The primers should be designed in the conserved region...
Embodiment 2
[0114] The optimization of embodiment 2 fluorescence quantitative PCR primer consumption
[0115] 1. The first optimization of the amount of fluorescent quantitative PCR primers
[0116] With diluted L1 (10 -4 ) and L2 (10 -5 ) positive control plasmid was used as a template for optimizing the amount of primers, and gradient optimization was performed on the upstream and downstream amounts of primers respectively. The concentration of the primer working solution was 10 pmol / μL. Polymerase (5U / μL) 0.6 μL, real-time fluorescent PCR buffer (2×) 12.5 μL (2.5 μL of ThermoFisher’s 10× buffer 2.5 μL, 2 μL of 10 mM dNTP and 10000×SYBRgreen I 0.0025 μL and 8 μL of DEPC water mixture), supplemented with sterile water to 25 μL. The PCR reaction conditions are: 95°C for 3min; 95°C for 10sec, 55°C for 20sec, 72°C for 20sec, 75°C for 5sec, and read plate for a total of 45 cycles. The results are shown in Table 3. It can be analyzed from the results that the PCR primers can work normally...
Embodiment 3
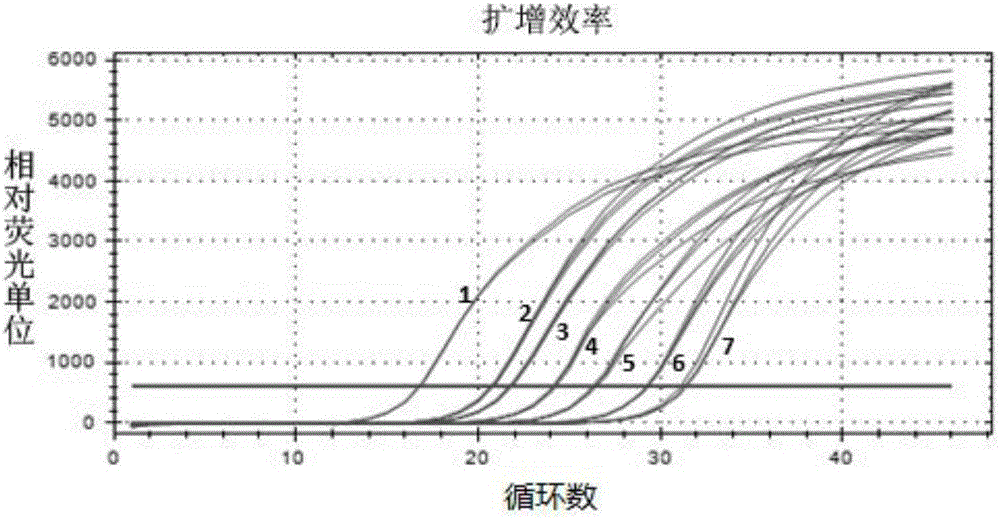
[0127] Example 3 Koi Herpesvirus Type III TK Gene Fluorescent Quantitative PCR Detection
[0128] 1. Establishment of standard curve
[0129] The positive control plasmid DNA was used as a template for fluorescent quantitative PCR detection to establish a standard curve. The specific operation is as follows: the plasmid DNA was serially diluted 10 times to I: 1.0×10 6 copies / μL; II: 1.0×10 5 copies / μL; III: 1.0×10 4 Copy / μL; Ⅳ: 1.0×10 3 Copy / μL; V: 1.0×10 2 Copy / μL; VI: 1.0x10 1 Copy / μL; VII: 1.0×10 0 copies / μL. Each dilution was repeated 3 times in parallel. Standard product detection PCR reaction system is: upstream primer 2.0 μL (10 pmol / μL), downstream primer 2.0 μL (10 pmol / μL), plasmid DNA 5 μL, Taq DNA polymerase (5U / μL) 0.6 μL, real-time fluorescent PCR buffer ( 2×) 12.5 μL (prepared with 2.5 μL of 10× buffer purchased from ThermoFisher, 2 μL of 10 mM dNTP, and a mixture of 0.0025 μL of 10000×SYBR green I and 8 μL of DEPC water), supplemented with sterile wate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com