Recombinant nucleic acid fragment RecCR020127 and detection method thereof
A technology for recombining nucleic acids and fragments, applied in the field of recombinant nucleic acid fragments and their detection, can solve the problems of polluting the environment, increasing production costs, and enhancing the drug resistance of N. lugens
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1 Breeding of recombinant plants introduced into the brown planthopper resistant genome fragment
[0045] The materials used in this embodiment are rice'Shengshi B'and rice'Hua 2048B'.
[0046] Rice'Hua 2048B' has good resistance to brown planthopper, which may be QBph3 (Hu et al., Molecular Breeding. 2015, 35: 3) and Bph14 (Du et al., PNAS. 2009, 106(52): 22163) on chromosome 3. -22168) played a key role in the brown planthopper resistance of this material.
[0047] In the process of selection of recombinant plants, molecular markers were used for prospect selection of recombinant plants, and the used prospect selection molecular markers were screened. The molecular markers used are partly derived from the website http: / / www.gramene.org / and partly designed by ourselves. The design method is to download the aforementioned segment DNA sequence according to the 6.1 edition of the MSU / TIGR annotation of the rice Nipponbare genome. Use SSRLocator software to scan the ...
Embodiment 2
[0056] Example 2 Determination of homologous recombination fragments after introduction of brown planthopper resistance genome fragments
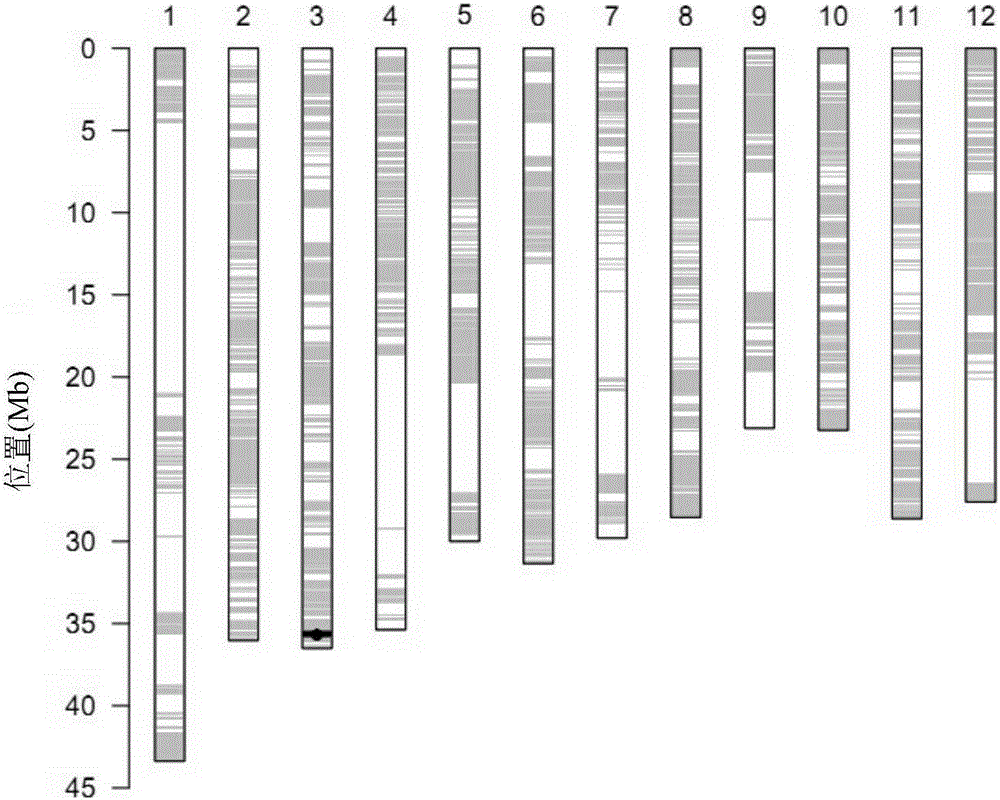
[0057] In order to determine the size of the introduced brown planthopper-resistant genome fragment, the homozygous single strain of the introduced fragment of'Shengshi B'was sequenced for the homologous recombination fragments on both sides of the target genome fragment. The recombinant nucleic acid fragment of the brown planthopper resistant genome contained in CR020127 was named RecCR020127.
[0058] It is preliminarily determined by the detection result of rice whole genome breeding chip RICE60K that RecCR020127 is located between the two SNP markers F0335474244AG and R0335762765GA.
[0059] At the same time, the Miseq sequencing technology was used to perform whole genome sequencing on three samples of'Shengshi B','Hua 2048B' and CR020127. Use TruSeq Nano DNA LT Kit (Illumina) kit for library establishment, use Library Quantification Kit–Un...
Embodiment 3
[0069] Example 3 Resistance identification of ‘Shengshi B’ after introducing the brown planthopper resistant genome fragment
[0070] In order to identify the resistance effect, the new line CR020127 selected in this application, the recipient parent'Shengshi B', the donor parent'Hua 2048B' (as a positive control), and the highly susceptible brown planthopper variety'Taichung Zailai 1'(as Negative control) for indoor resistance identification of brown planthopper, the identification method is as follows.
[0071] Soak each material in the room, accelerating germination, and then sown in a plastic basin with a grid line. Each material is divided into 3 rows and a total of 45 plants are sown. When the two leaves and one heart period are reached, 10 plants and 30 plants in each row are kept healthy Consistent plants are used for infestation. The source of the insects comes from the brown planthoppers collected from the field and reared indoors. Take 2-3 instar nymphs on the plants...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com