SNP molecular marker in close linkage with bacterial blight resistance gene Xa7 and applications of SNP molecular marker
A bacterial blight resistance and molecular marker technology, which can be used in recombinant DNA technology, microbial assay/test, DNA/RNA fragments, etc. Design gene function markers and other issues to achieve the effect of facilitating high-throughput rapid detection, eliminating aerosol pollution, and accurate detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] This example is used to illustrate the application of the molecular marker provided by the present invention in the detection of bacterial blight resistance gene Xa7, and the specific steps are as follows:
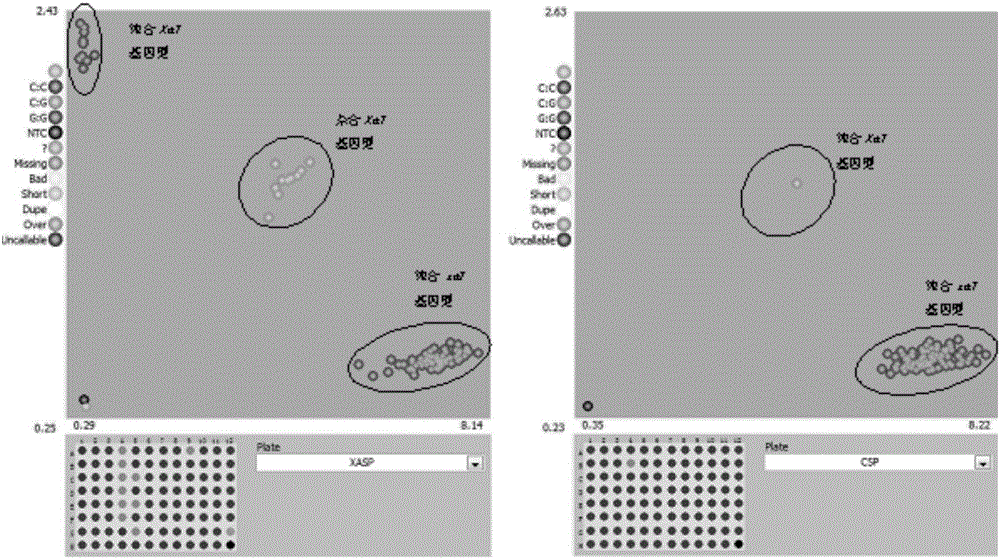
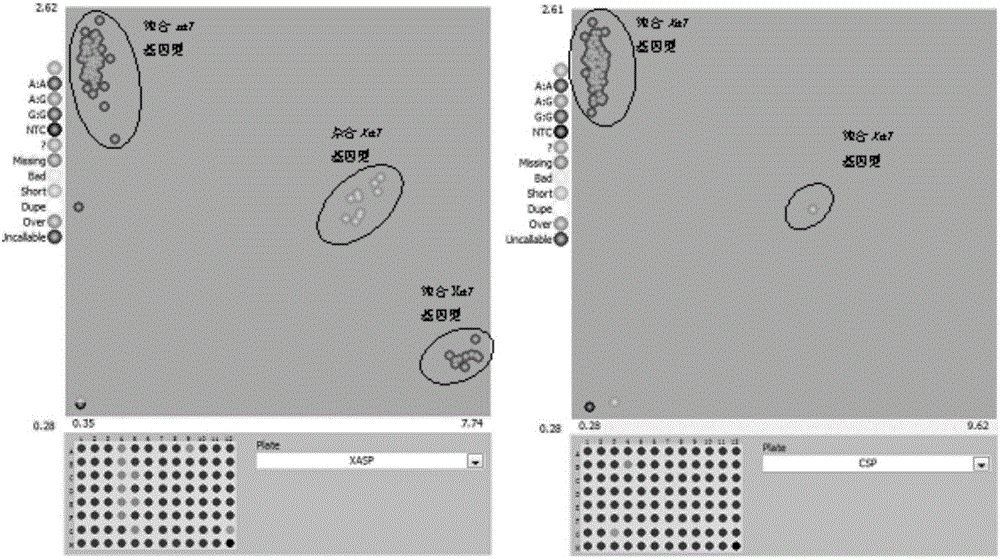
[0035] 1. According to the molecular markers described in the present invention, design primer combinations that can be used for high-throughput detection of Xa7 resistance gene in rice materials by using KASP reaction, as shown in Table 1.
[0036] 2. Genomic DNA was extracted from rice leaves by simplified CTAB method.
[0037] ①Put the sample into a 2.0mL Tube, add two steel balls and 750μL CTAB solution in advance, shake the homogenized sample for 1.5min;
[0038] ②Concussion heating at 65℃ for 0.5-1h;
[0039] ③ Cool to room temperature, add 750mL chloroform:isoamyl alcohol (24:1) solution in the fume hood and mix well;
[0040] ④ Centrifuge at 12000rmp for 10min, take about 500mL supernatant and transfer it to a new 1.5mL centrifuge tube;
[0041] ⑤ Add an ...
Embodiment 2
[0056] 1. Genetic position verification of molecular markers
[0057] Nine genome-wide markers with polymorphisms in the parents and two markers linked to Xa7 in the present invention were used to verify the genetic position. The two molecular markers linked to Xa7 in the present invention were both mapped to chromosome 6 82.7 cM position, such as image 3 shown.
[0058] 2. Phenotypic verification of molecular markers
[0059] The genetic phenotype of the two Xa7-linked markers in the present invention was verified by using 120 F2 hybrid populations of the donor parent Hua 1228S and the recurrent parent R608. At the rice booting stage, 120 individual plants of the F2 population and 2 parental varieties were inoculated with the Guangdong bacterial blight strain GD1358, and the disease was investigated 21 days later. The consistency between the phenotype data and the genotype was more than 90%, which verified the method of the present invention again. Feasibility and accurac...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com