Detection method for salmonella enteritidis and detection kit
A detection kit and Salmonella detection technology, which is applied in biochemical equipment and methods, microbial measurement/inspection, and resistance to vector-borne diseases, etc. It can solve the problems that the detection sensitivity needs to be further improved, it is difficult to promote on a large scale, and it is difficult to detect directly , to achieve the effect of rapid response, low cost and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
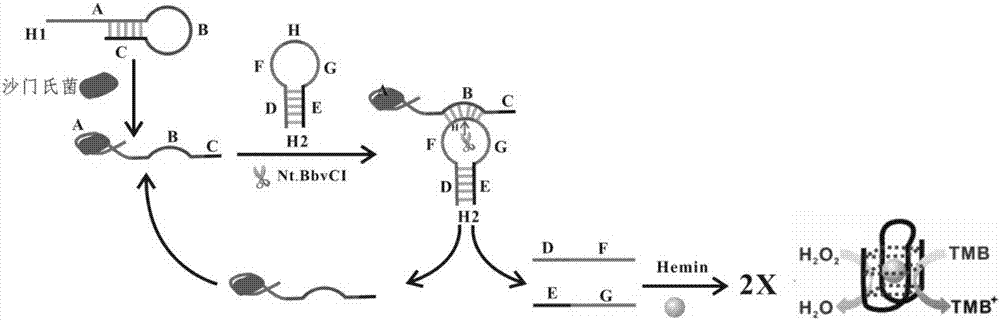
[0073] A detection method for salmonella, carried out according to the following steps:
[0074] (1) Use Tris-HCl buffer (10 mM, pH 7.9, containing 50 mM NaCl, 10 mM MgCl 2 and 100 μg / ml BSA) to dissolve nucleic acid H1 and H2 respectively. Mix 1 M H1 with Salmonella and react at room temperature for 30 minutes.
[0075] (2) Add 1 M H2, mix well, and react at room temperature for 30 minutes.
[0076] (3) Add 25 U of endonuclease Nt.BbvCI, mix thoroughly, and react at room temperature for 90 minutes.
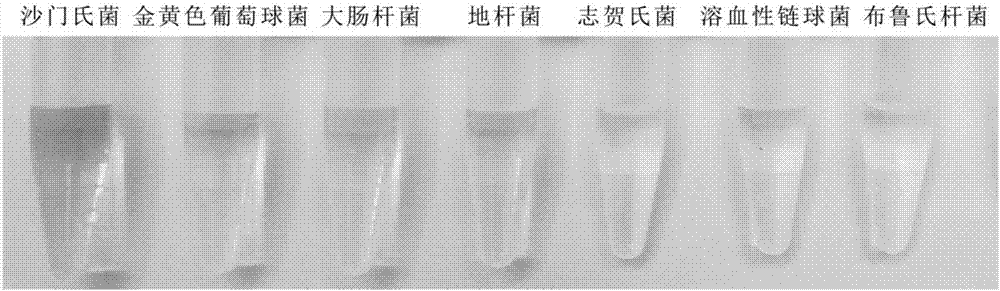
[0077] (4) Add 0.3 M Hemin (hemin) and react at room temperature for 30 minutes. Take out 50 L of reaction solution and add it to 950 L of chromogenic buffer system (containing 26.6 mM citric acid, 51.4 mM disodium hydrogen phosphate, 25 mM KCl, 10 L of 0.5% TMB, 20 L of 30% H 2 o 2 , pH=5.0), react at room temperature for 15 minutes, and observe the color change. When Salmonella is present, the solution turns blue, and when there is no Salmonella, the solution is colorless....
Embodiment 2
[0079] A Salmonella detection kit includes the following components:
[0080] (1) Nucleic acids H1 and H2, the sequences of which are as follows:
[0081] Nucleic acid sequence H1: 3'-TGATGGCTGTAGTGTTTCCGGGTTATCACTAGTTTGGTGGCACCAATGTCAGTCTCCTC (A)-TTCGGAGTCGCTA (B)-GAGGAGAC (C)-5' (SEQ ID NO: 1);
[0082] Nucleic acid sequence H2: 5'-AGTACTAG (D)-GGGTAGGGCGGGTTGGG (F)-CC↓TCAGC (H)-GGGTAGGGCGGGTTGGG (G)-CTAGTACT (E)-3' (SEQ ID NO: 2); (the arrow indicates the cleavage site );
[0083] (2) Endonuclease Nt.BbvCI;
[0084] (3) Hybridization buffer containing 10 mM Tris-HCl, pH 7.9, containing 50 mM NaCl, 10 mM MgCl 2 and 100 μg / ml BSA;
[0085] (4) Hemin;
[0086] (5) Chromogenic buffer system, including 26.6 mM citric acid, 51.4 mM disodium hydrogen phosphate, 25 mM KCl, 10 L 0.5% TMB, 20 L 30% H 2 o 2 , pH=5.0.
Embodiment 3
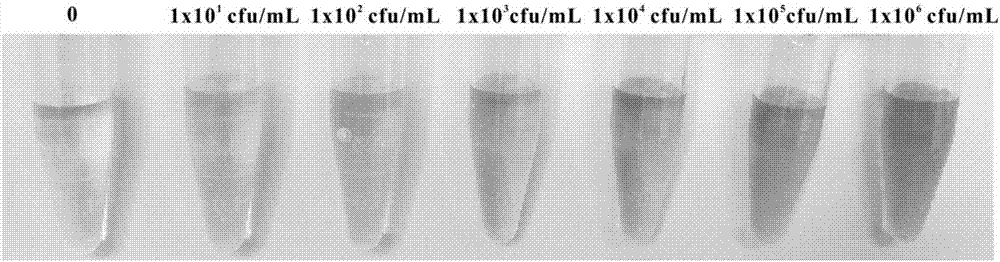
[0088] Detection of different concentrations of Salmonella:
[0089] Prepare Salmonella standard solution, the concentration is 1x10 1 cfu / mL, 1x10 2 cfu / mL, 1x10 3 cfu / mL, 1x10 4 cfu / mL, 1x10 5 cfu / mL, 1x10 6 cfu / mL Store at 4°C. The Salmonella solution of different concentrations is added respectively in the reaction system described in embodiment 1, observes experimental result after fully reacting, as figure 2 As shown, 10 cfu / mL of Salmonella can produce a clear blue change, indicating that its detection limit is 10 cfu / mL. As the concentration of Salmonella increased, the color also increased and gradually became saturated.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com