Adjustable luciferase segmentation fusion protein and preparing method and application thereof
A luciferase and fusion protein technology, applied in the field of molecular biology, can solve the problems of poor real-time performance and differences of dual fluorescence complementary technology, which can only be detected successfully in a few minutes or even a few hours
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Preparation of Gausssia luciferase fusion fragment
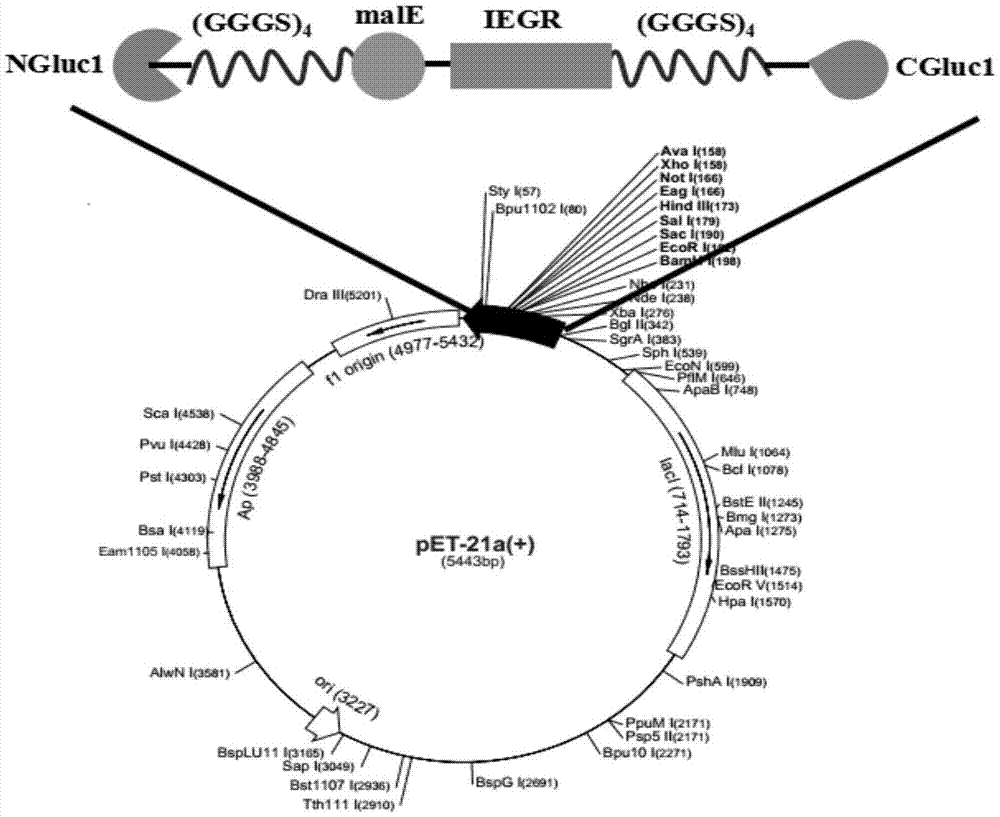
[0048] (1) Select the expression vector of the luciferase fusion gene as pET21a, and then select a suitable insertion site for luciferase, and finally choose to insert an exogenous fragment between Arg93 and Cys94 to form the N fragment NGluc1 (1~ 93aa) and C fragment CGluc1 (94~185aa);
[0049] (2) Using the recombinant vector pET28a-Gluc1 as a template, when designing and amplifying the NGluc1 fragment, introduce BamH Ⅰ and EcoR Ⅰ restriction sites at the 5' end of the primer, and when designing and amplifying the CGluc1 fragment, introduce HindⅢ at the 5' end of the primer and Not I restriction sites, and are designed to introduce flexible peptide chains (GGGGS) 4 To reduce the steric hindrance caused by specific enzyme cleavage;
[0050] The designed primer sequences are as follows:
[0051] Gluc1 upstream primer (BamH Ⅰ):
[0052] 5'-CGCGGATCCATGAAGCCCACCGAGAACAACGAAG-3';
[0053] NGluc1 downstream...
Embodiment 2
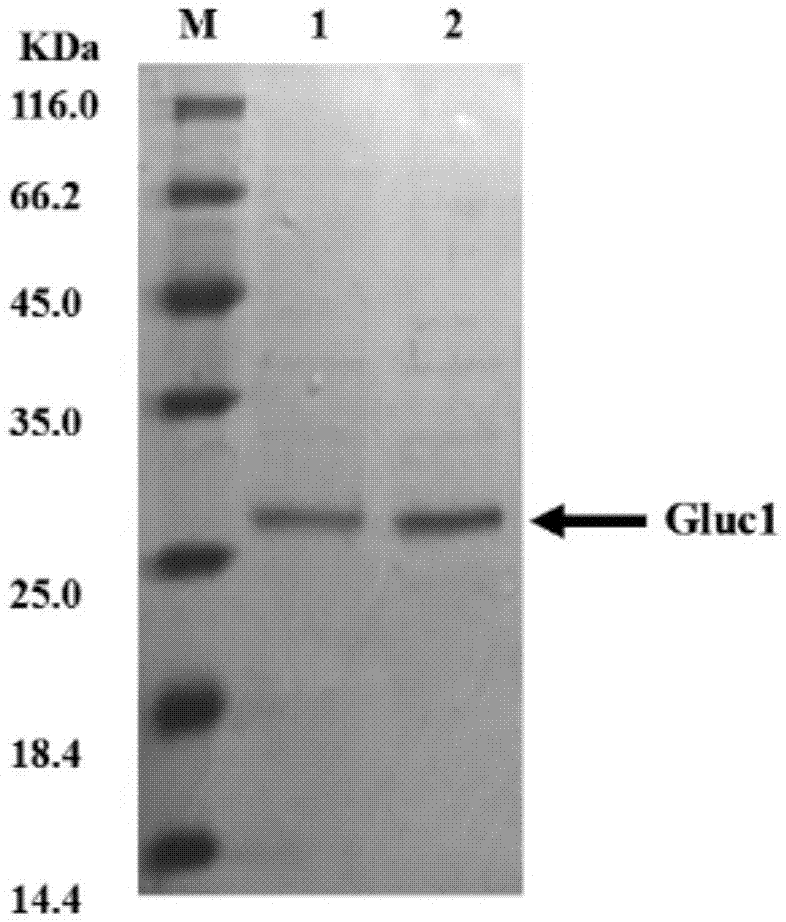
[0069] Embodiment 2 Expression and purification of luciferase fusion body and Gaussia luciferase
[0070] (1) Strain activation: inoculate frozen glycerol bacteria pET21a-NGluc1-malE-CGluc1 and pET28a-Gluc1 into LB liquid medium (containing 100 μg / mL ampicillin) at 37°C and 225 rpm to activate overnight strains;
[0071] (2) Strain fermentation: Inoculate the activated bacterial liquid into fresh LB liquid medium (containing 100 μg / mL ampicillin) at a ratio of 1:100 and cultivate to OD 600 When it reaches 0.4-0.6, it is the logarithmic phase of the growth of the strain;
[0072] (3) Induced expression: add inducer isopropyl-β-D-1-thiogalactoside IPTG (final concentration is 1mM) to induce high-speed expression of luciferase fusion at 28°C, centrifuge at 11000rpm at 4°C Collect the thalli within 10min, discard the supernatant LB medium, add a certain amount of binding buffer (20mM Na 3 PO 4 , 0.5M NaCl, 40mM imidazole, pH 7.4) suspended bacteria;
[0073] (4) Ultrasonic dis...
Embodiment 3
[0075] Example 3 Construction of the dynamic interaction model of luciferase fusion protein
[0076] (1) Realize the ON mode of protein dynamic interaction
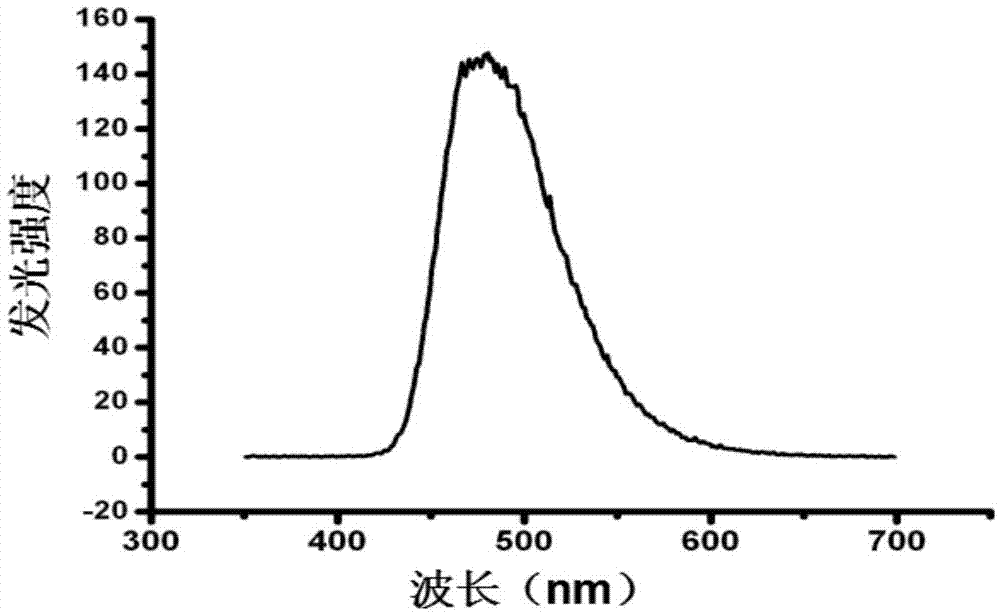
[0077] Add 1×PBS (pH7.4) buffer solution to a 96-well white microtiter plate, add luciferase fusion protein diluted to a certain concentration with buffer solution, and quickly add intestinal Coelenterazine (Coelenterazine) substrate, to ensure that the total volume of the system is 200 μL, after thorough mixing, immediately use a microplate reader to detect the bioluminescent intensity in the system (the emission spectrum of the fluorescein surface is shown in image 3 As shown, the detection wavelength is 480-500nm), and the ON mode of the protein dynamic interaction model is realized. The model was evaluated with Gaussia luciferase as a control group;
[0078] (2) Realize the OFF mode of protein dynamic interaction
[0079] Add a certain concentration of cutting enzyme Factor Xa Protease to the reaction system, mix ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com