SNP (single nucleotide polymorphism) marks of Yuxi fat-tailed sheep as well as screening method and application of SNP marks
A screening method and labeling technology, which can be used in biochemical equipment and methods, recombinant DNA technology, DNA/RNA fragments, etc. , the effect of no interference between sites
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Determination of Yuxi fat-tailed sheep breeds by pyrophosphoric acid method
[0036] (1) Reagents: GoTaqMaster Mix solution produced by Promega Company; specific amplification primers and sequencing primers synthesized by Shanghai Sangon; Sepharose Bead produced by Biotage Company.
[0037] (2) Amplification reaction system and amplification program: the total volume of the amplification reaction is 50 μL, and its various components are: 2×GoTaqMaster Mix 25 μL, 10 μmol / L primers 1 μL each, DNA template 2 μL for the test variety and the control variety (Cucumber seed DNA), make up to 50 μL with sterilized deionized water; reaction program: pre-denaturation at 95°C for 10 minutes, denaturation at 94°C for 30 seconds, annealing at 50°C for 30 seconds, extension at 72°C for 45 seconds, 50 cycles, and finally extension at 72°C for 7 minutes , stored at 4°C.
[0038] (3) Sequencing reaction system: Add 47 μL Binding buffer and 3 μL Sepharosebeads to 50 μL PCR prod...
Embodiment 2
[0041] Example 2PCR amplification method for determination of Yuxi fat-tailed sheep breeds
[0042] Randomly select 24 Altay sheep, 24 Bayinbulak sheep and 29 Yuxi fat-tailed sheep as sample individuals, collect the blood of these sheep, anticoagulate with heparin, and extract genomic DNA with Tiangen blood genome kit, 0.8% Agarose gel electrophoresis detection, nucleic acid protein quantification instrument to measure its concentration and purity, diluted to 100ng / ul, and stored for future use. Use the primers listed in Table 2 to perform PCR amplification using the genomic DNA of the sample to be tested as a template. The PCR amplification reaction system is: add 0.4 μL of Taq DNA polymerase (5U / μL), 5 μL of 10×PCR Buffer, and 4 μL of MgCl2 (25mmol / μL), 1μL of Forward primer (10pmol / μL), 1μL of Reverse primer (10pmol / μL), 4μL of dNTP (2.5mmol / μL), 2μL of template DNA and 32.6μL of ddH2O, a total of 50μL.
[0043] PCR products were recovered and purified using an agarose gel...
Embodiment 3
[0044] Example 3 Determination of Yuxi fat-tailed sheep breeds by gene chip method
[0045] Step 1, taking experimental animals, taking blood from ear veins, and extracting genomic DNA from the blood samples.
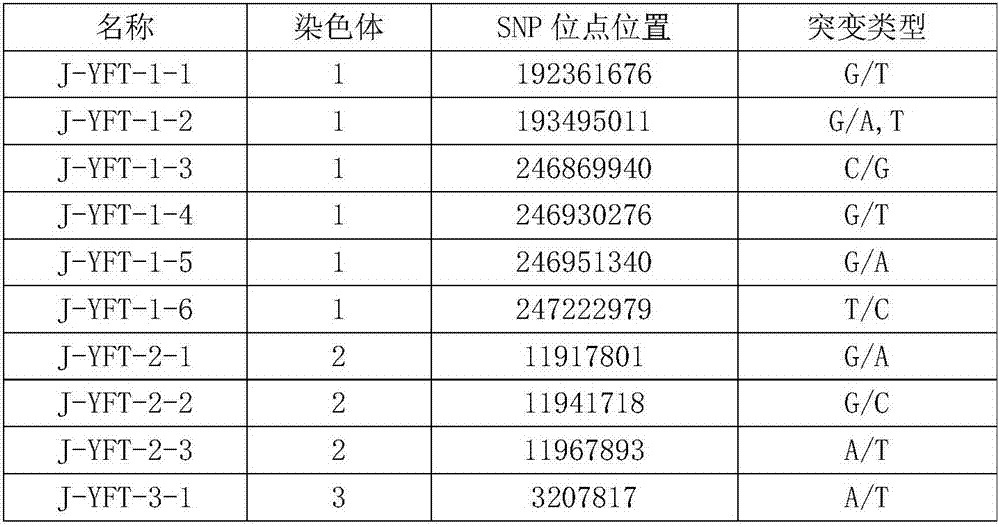
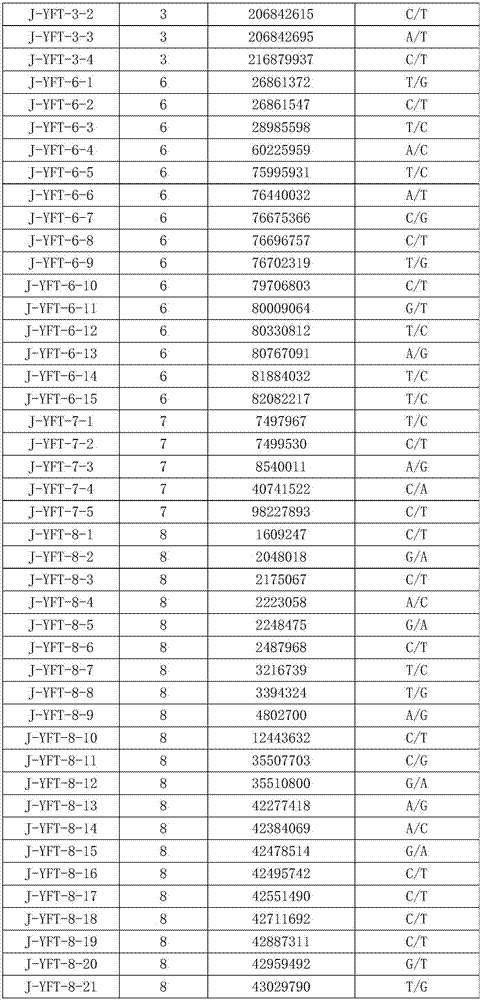
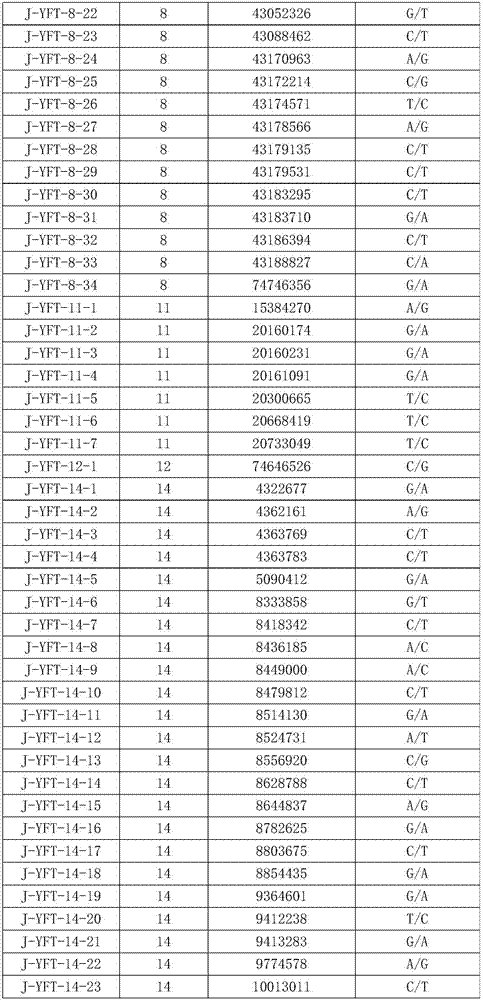
[0046] Step 2. Take the genomic DNA obtained in step 1 and hybridize it with the nucleic acid chip with inherent 149 probes (30 probes are the single-stranded DNA molecules shown in sequence 1 to sequence 30 in Sequence Table 2).
[0047] Step 3. After step 2 is completed, the ends of each point in the nucleic acid chip are extended, so as to obtain the genotypes of 149 SNP sites in the genomic DNA.
[0048] Step 4, taking the genomic DNA obtained in step 1, performing whole genome sequencing, and obtaining the genotypes of 149 SNP sites in the genomic DNA. The result shows that the result of step 3 is exactly the same as the result of step 4. Among the 149 SNP sites of 48 experimental animals of Yuxi fat-tailed sheep, at least 99-136 sites could be matched. The matc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com