Dracaena cambodiana chalcone isomerase DcCHIL1, encoding gene thereof and application
A chalcone isomerase and gene technology, applied in the field of plant biology, can solve problems such as inability to increase the content of flavonoids, and achieve the effects of great economic value, broad application prospects, and yield improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Cloning of Chalcone Isomerase Gene DcCHIL1 from Dracaena hainanensis
[0030] Through the analysis of the sequencing results of the stem transcriptome of Dracaena hainanensis in the early stage, the specific primers P1 forward primer 5'-ATGGGCTCTGAGATAGTGATGGTG-3'(SEQ ID NO:3) and P2 reverse primer 5'-GCAGCAGATAACATAGTCCCAAG-3'( SEQ ID NO: 4), the CDS sequence of DcCHIL1 gene was amplified by PCR using the first-strand cDNA of the stem of Dracaena hainan as a template, and the amplification conditions were: pre-denaturation at 94°C for 5min; denaturation at 94°C for 30sec, annealing at 55°C 30sec, 70°C extension for 1min, a total of 35 cycles; 70°C extension for 8min. The amplified PCR product was connected to the pMD18-T vector, transformed into Escherichia coli competent cells, positive clones were screened and sequenced, and the full-length coding sequence 642bp of the chalcone isomerase gene DcCHIL1 of Dracaena hainanensis was obtained.
[0031] The above...
Embodiment 2
[0032] Example 2 Sequence analysis of the chalcone isomerase gene DcCHIL1 from Dracaena hainanensis
[0033] The full-length open reading frame of the chalcone isomerase gene DcCHIL1 of Dracaena hainanensis is 642bp, and the detailed sequence is shown in SEQ ID NO:1. The amino acid sequence of DcCHIL1 was deduced according to the sequence of the open reading frame, with a total of 213 amino acid residues (see SEQ ID NO: 2 for the detailed sequence), a molecular weight of 23.8 kDa, and a theoretical isoelectric point of 4.97.
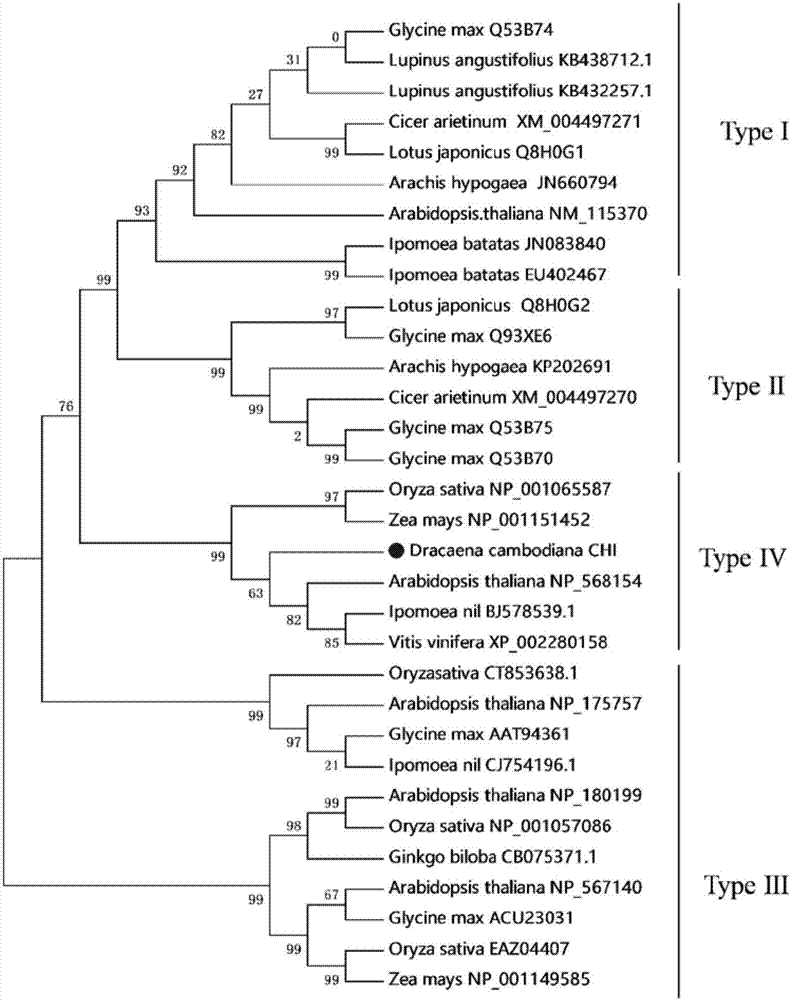
[0034] 31 different types of CHI sequences were selected from different plants such as Arabidopsis thaliana and corn (for details of plant species and CHI protein sequence numbers, see figure 1 ) as a template to analyze the phylogenetic status of DcCHIL1. Use MEGA7 software to perform multiple sequence alignment on these 32 CHI sequences, and construct a phylogenetic tree (such as figure 1 ). It can be clearly seen from the figure that these CHIs can...
Embodiment 3
[0035] Example 3 Construction of PET28a-DcCHIL1 recombinant expression vector
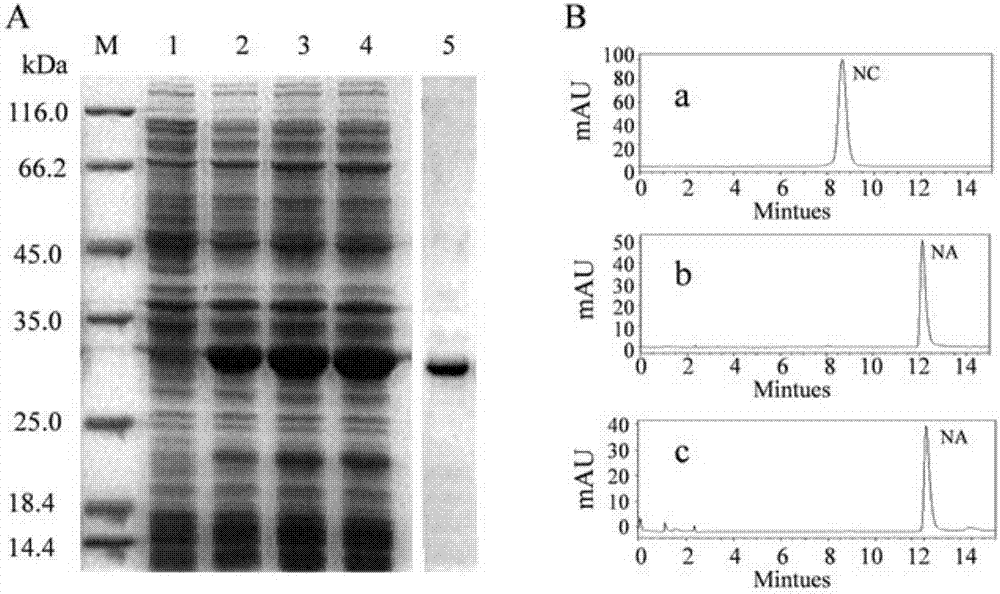
[0036]Using Premier 5 software to analyze the restriction endonuclease cutting sites of DcCHIL1 coding region of Dracaena hainanensis chalcone isomerase gene, the two restriction endonucleases used to construct the vector were determined to be EcoRI and SalI. The primers PE1: 5′-GCGAATTCATGGGCTCTGAGATAGTGATGGTG-3′ (forward primer, marked as the EcoRI restriction site) and PE2: 5′-GCGTCGACAGCAGCAGATAACATAGTCCC-3′( Reverse primer, marked as the SalI restriction site), and pMD18-DcCHIL1 (prepared in Example 1) was used as a template for PCR amplification. The amplification conditions were: 94°C pre-denaturation for 5min; 94°C denaturation for 30sec, 58°C Anneal for 30 sec, extend at 70°C for 1 min, total 35 cycles; extend at 70°C for 8 min. The amplified PCR product was connected to the pMD18-T vector to obtain the pMD18-E DcCHIL1 recombinant plasmid, and the correctness of the target sequence was co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com