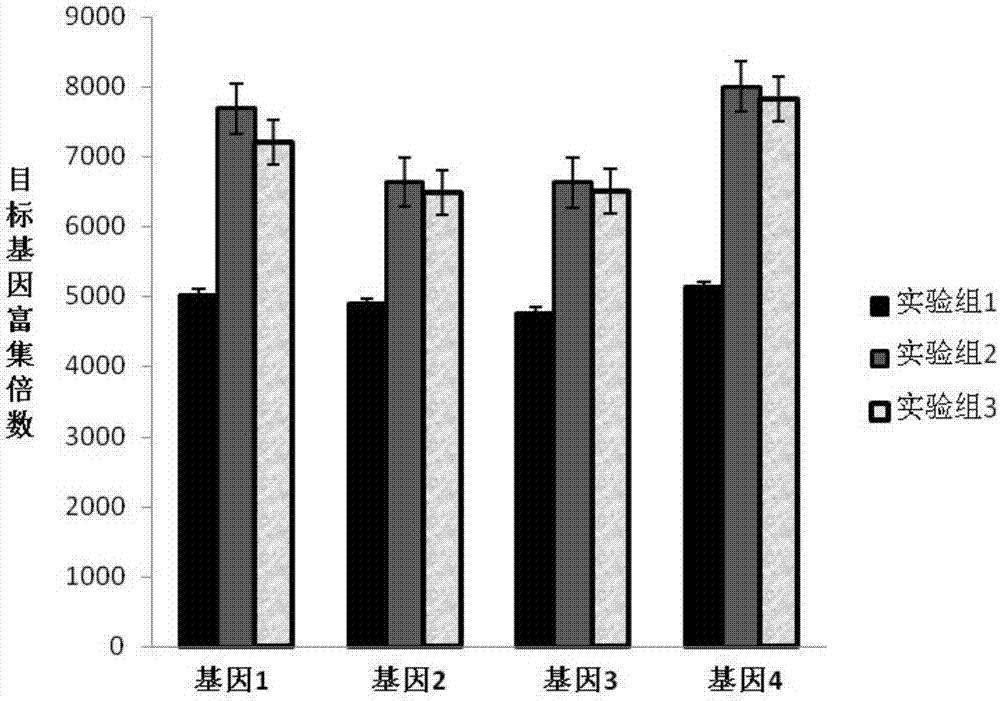
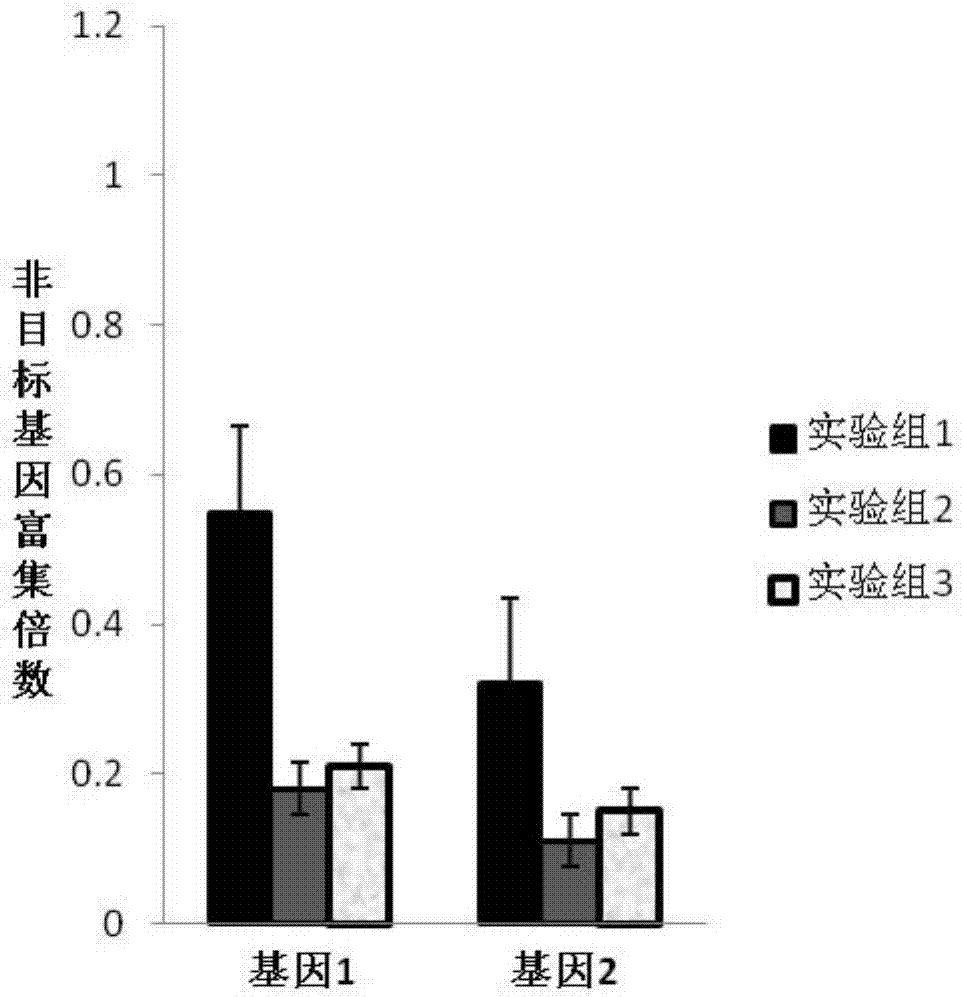
Liquid-phase hybrid capture kit based on double-stranded probe, washing kit and application of kits
A technology of hybrid capture and kit, which is applied in the field of molecular biology, can solve the problems of high synthesis cost of molecular probes, insufficient detection sensitivity, and difficult region amplification, etc., so as to improve target enrichment efficiency, reduce costs, and achieve high efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation Embodiment 1
[0039] Preparation of Example 1 Capture Solution Preparation
[0040] The solution consists of sodium dihydrogen phosphate (NaH 2 PO 4 ) buffer, sodium citrate buffer (SSC), EDTA
[0041] (EDTA), deionized formamide, Denhardt (Denhardt) solution, Carrier RNA and dextran sulfate
[0042] made. First prepare a 2×concentration solution (without deionized formamide, deionized formamide liquid is added separately to capture
[0043] solution), it can be stored for a long time at -20°C, and the specific formula is as follows:
[0044] 100mM NaH 2 PO 4
[0045]4×SSC
[0046] 2mM EDTA
[0047] 2 x Denhard's solution
[0048] 2mg / ml Carrier RNA
[0049] 20w / w% dextran sulfate
[0050] Solution pH 7.0
[0051] The capture solution includes 2×concentration solution and 100% formamide liquid, which are mixed to form a hybridization capture solution working solution, and the volume mixing ratio is shown in Table 1 below.
[0052] Table 1
[0053]
preparation Embodiment 2
[0054] Preparation Example 2 Preparation of Washing Solution Kit
[0055] The washing solution kit contains 5 washing buffer components, which can be stored for a long time at -20°C. The specific formula is as follows:
[0056] Stringent wash solution: 2 x SSC solution, 30 v / v % deionized formamide.
[0057] Wash solution I: 2 x SSC solution, 0.1% sodium dodecyl sulfate (SDS).
[0058] Wash solution II: 2x SSC solution.
[0059] Wash solution III: 0.2x SSC solution.
[0060] Biotin affinity beads washing solution: 10mM Tris-HCl, pH 7.5; 1mM EDTA; 2M NaCl.
Embodiment 1
[0061] Example 1 Application of capture solution and washing solution kit in hybrid capture enrichment
[0062] Specifically, the entire experimental process includes: firstly, the extracted sample DNA is ultrasonically broken into DNA fragments of 200-250bp (but not limited to the ultrasonic breaking method), and the DNA The fragments are connected to the index adapter; then the constructed capture library is used to hybridize with the DNA fragments containing the target gene region to be captured, the DNA fragments in the target region are captured, the enrichment effect of the DNA fragments in the target region is evaluated, and these DNA fragments are sequenced. The sample detection results were obtained after bioinformatics data analysis.
[0063] 1. Sample genomic DNA extraction and library preparation
[0064] The extraction method of the genomic DNA of the samples refers to the QIAamp DNA Blood Mini Kit kit of Qiagen Company, and the human whole blood genomic samples ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com