Intelligent thermostatic amplification technology based salmonella detection primer group, kit and method
A constant temperature amplification, Salmonella technology, applied in the field of molecular biology, can solve the problems of high technical level requirements of testing personnel, difficulty in realizing rapid testing promotion, expensive testing instruments, etc., and achieves overcoming genetic polymorphism detection and accurate genetic polymorphism Sex, the effect of short detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Establishment of Salmonella Intelligent Constant Temperature Amplification Detection Kit
[0035] Intelligent constant temperature amplification detection kit for Salmonella, including intelligent constant temperature amplification primer set, intelligent constant temperature amplification reaction solution, Bst DNA polymerase, mismatch binding protein Tap Muts, positive control and negative control, fluorescent chromogenic reagent.
[0036](1) Design of intelligent constant temperature amplification primers: design primers with the Salmonella invA gene as the target gene. The primer sequences are listed in Table 1.
[0037] Table 1 Primer sequence list
[0038] Primer name
Primer sequence (5'-3')
FP
TTTATATATATATAAAGTTATTGGCGATAGCC (SEQ ID NO: 1)
TP
AGCCTGGCGGTTGGACCACGGTGACAATAG (SEQ ID NO: 2)
BP
GGTTTTGTTGTCTT (SEQ ID NO: 3)
OP1
CGCCACGTTCGGGCAATT (SEQ ID NO: 4)
OP2
TTTGGTAATAACGATAAA (SEQ...
Embodiment 2
[0042] Example 2 uses the ESE-Quant tube scanner to establish a Salmonella detection method:
[0043] The method utilizing the test kit of embodiment 1 to detect Salmonella comprises the steps:
[0044] (1) Extraction of DNA from the sample to be tested: extract DNA from the sample using a bacterial DNA extraction kit;
[0045] (2) Constant temperature gene amplification reaction: 25 μL reaction system contains 1.6 μmol / L of TP and FP primers, 0.8 μmol / L of BP primers, 0.5 μmol / L of OP1 and OP2 primers, 0.5 mmol / L of dNTPs, and 0.6 μmol / L of betaine mol / L, (NH4)2SO4mmol / L, MgCl2 2.5mmol / L, KCl 10mmol / L, MgSO42mmol / L, Green I 0.5μL, Tris-HCl with pH 8.8 20mmol / L, 0.2% 20. 6U Bst DNA polymerase, 1 μg Tap Muts, 1 μL DNA template, make up to 25 μL with deionized water; set positive control and negative control; mix the prepared PCR tube, centrifuge, and react at 63 ° C for 40 min.
[0046] (3) Judgment of results: Place the reaction tube in the ESE-Tube Scanner for reaction, ...
Embodiment 3
[0047] Embodiment 3 specificity experiment
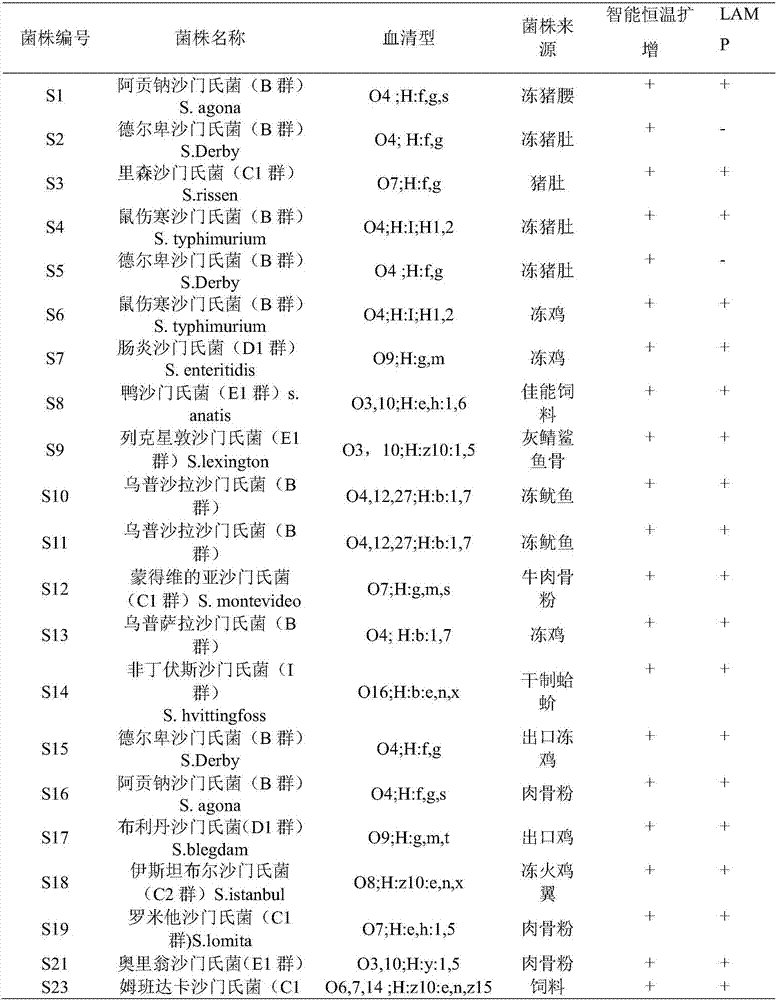
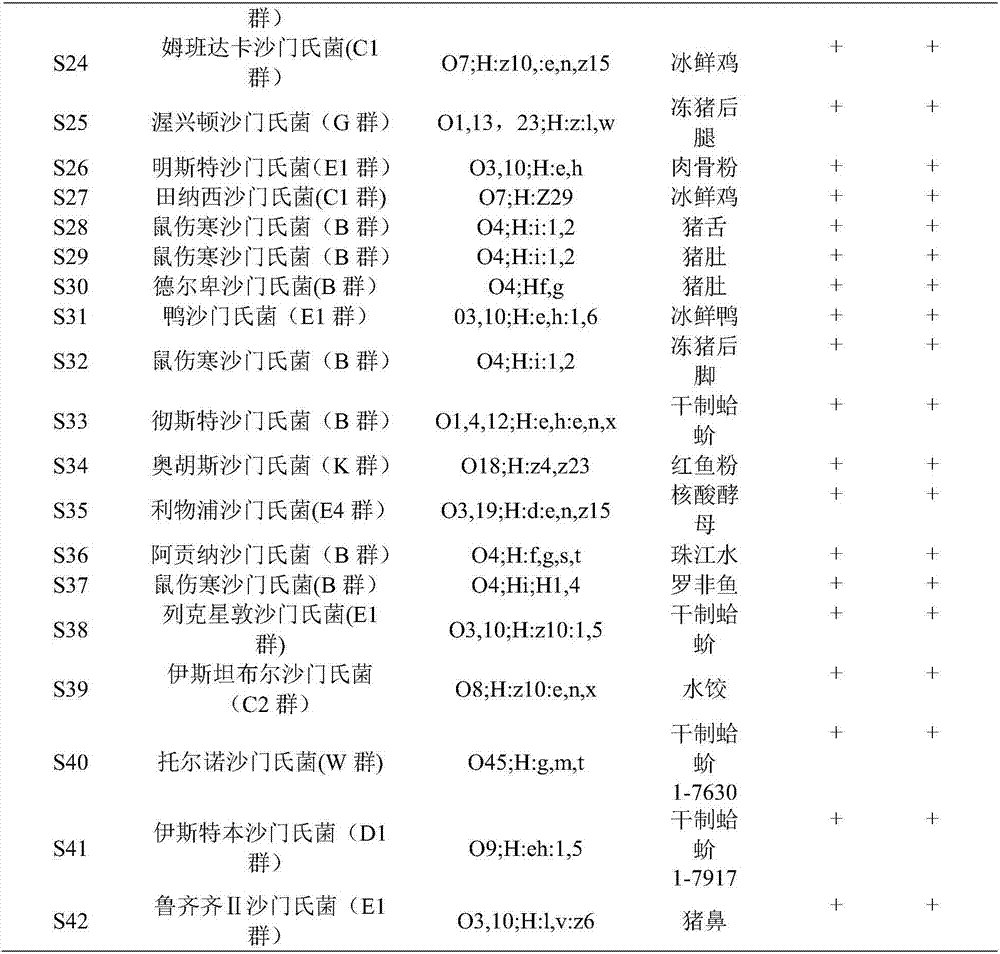
[0048] Using the method of Example 2 to separate and purify Listeria monocytogenes ATCC19115 / 413, Salmonella ATCC14028, Staphylococcus aureus ATCC6538, Escherichia coli O157ATCC43889, Enterobacter sakazakii ATCC29544, Pseudomonas aeruginosa ATCC9027, Escherichia coli ATCC25922, Escherichia coli CMCC44102, Enterobacter sakazakii ATCC12868, Vibrio parahaemolyticus ATCC17802, Vibrio vulnificus ATCC27562, Staphylococcus aureus CMCC26003, Shigella baumannii CMCC51346, Shigella sonnei NICP51334, Proteus vulgaris NICPB490059, The DNA of Proteus mirabilis HB7131, Klebsiella CMCC (B) 46117, and Micrococcus luteus CMCC28001 were detected.
[0049] The identification results showed that Salmonella ATCC14028 was amplified normally, and the DNA of other bacteria and the negative control were not amplified. showed good specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com