Micro-screening apparatus, process, and products
A microcavity and cavity technology, applied in measuring devices, library screening, biochemical equipment and methods, etc., can solve problems such as high cost materials and reagents, difficult instantaneous measurement of kinetic parameters, low throughput, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0247] Example 1: Microarray Single Cell Analysis and Laser Extraction
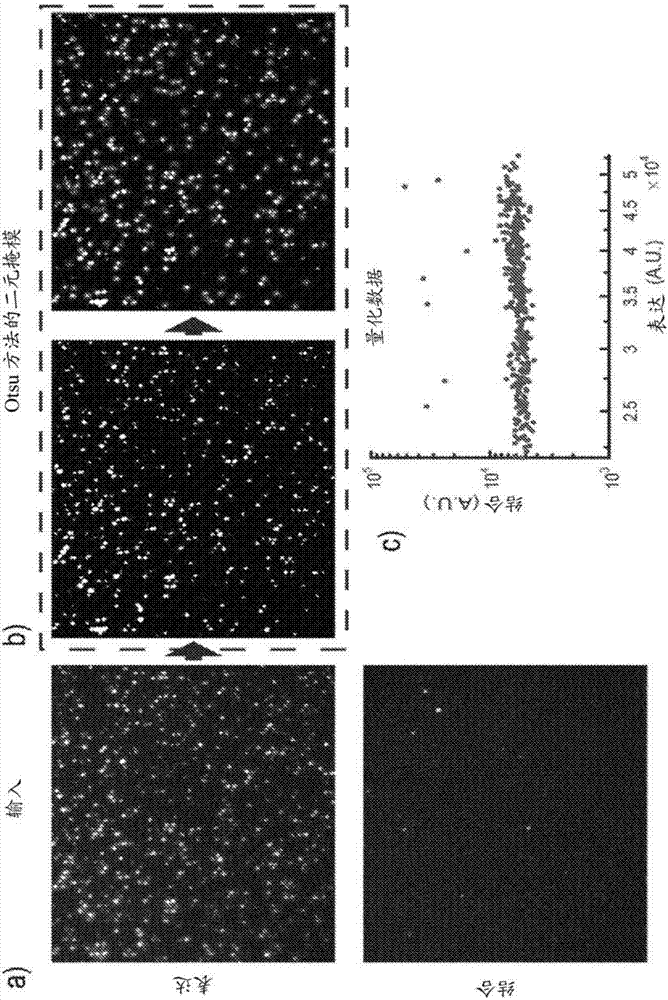
[0248] Figures 6A-6D depict the general concept and workflow of a microarray of one aspect of the present disclosure (referred to herein as "μSCALE"). Figure 6A shows the platform workflow. 1) A library of protein variants expressed in bacterial or yeast cells is mixed with opaque magnetic beads. 2) Pipette the mixture into the array at a concentration that results in an average single cell occupancy within the microchamber. Various biochemical assays are performed, with or without cell growth, using fluorescence as readout. 3) The array is imaged via fluorescence microscopy. 4) Quantify the fluorescence intensity of each microcavity and use a laser-based extraction method to isolate desired clones from the array, either as single cells or as bulk pools. 5) Culture the extracted cells in liquid or solid medium. 6) Cells are lysed and plasmids are recovered for characterization and / or to generate new ...
Embodiment 2
[0264] Example 2: High Throughput Screening of Protein Binding Interactions
[0265] Axl IgI mock library screening. will encode human Axl Ig1 (amino acid Ala 19 -Pro 131 ) and non-binding Ax1 variants (E59R, T77R) were cloned into the pCT yeast display plasmid between the NheI and BamHI restriction sites (Kariolis, M.S. et al., Nat. Chem. Biol. 1-10 (2014 )). Yeast surface display studies were performed by electroporation of plasmid DNA into Saccharomyces cerevisiae strain EBY100. Soluble Gas6 was recombinantly expressed in human embryonic kidney (HEK) cells using the FreeStyle Max 293Expression System (Invitrogen) and purified as previously described (Kariolis, M.S. et al, Nat. Chem. Biol. 1-10 (2014)). Natural scFv libraries displayed on the surface of yeast were previously described ( variant) (Deventer, J.A. Van & Wittrup, K.D., Yeast Surface Display for Antibody Isolation: Library Construction, Library Screening, and Affinity Maturation. 1131, 151-181 (Humana Pres...
Embodiment 3
[0281] Example 3: Affinity maturation of the Ax1 receptor: error-prone library sorting
[0282] Gas6 / Ax1 was used as a model system to isolate high affinity Ax1 variants that are potent inhibitors of tumor metastasis. The protein engineering strategy involved two iterations of library generation and screening: an initial library of randomly generated Axl mutations and a second library of recombinant Axl mutations derived from variants recovered from the first library.
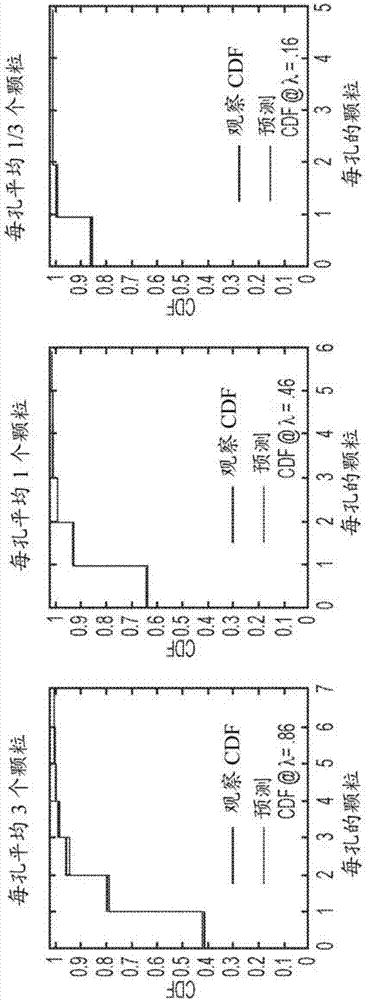
[0283] A round of MACS using Gas6-coated magnetic beads reduced the theoretical library size to 3.5 × 10 6 Clones were cloned, and arrays were loaded at a concentration that yielded an average of 4 cells per microtube.
[0284] The resulting pool of Ax1 variants was cultured, induced, and incubated with 100 pM Gas6 for 15 hours to allow equilibrium binding. The library was diluted to 12,600 cells / µL to a concentration of an average of 4 cells per 20 µm diameter capillary and imaged with epifluorescence in two...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com