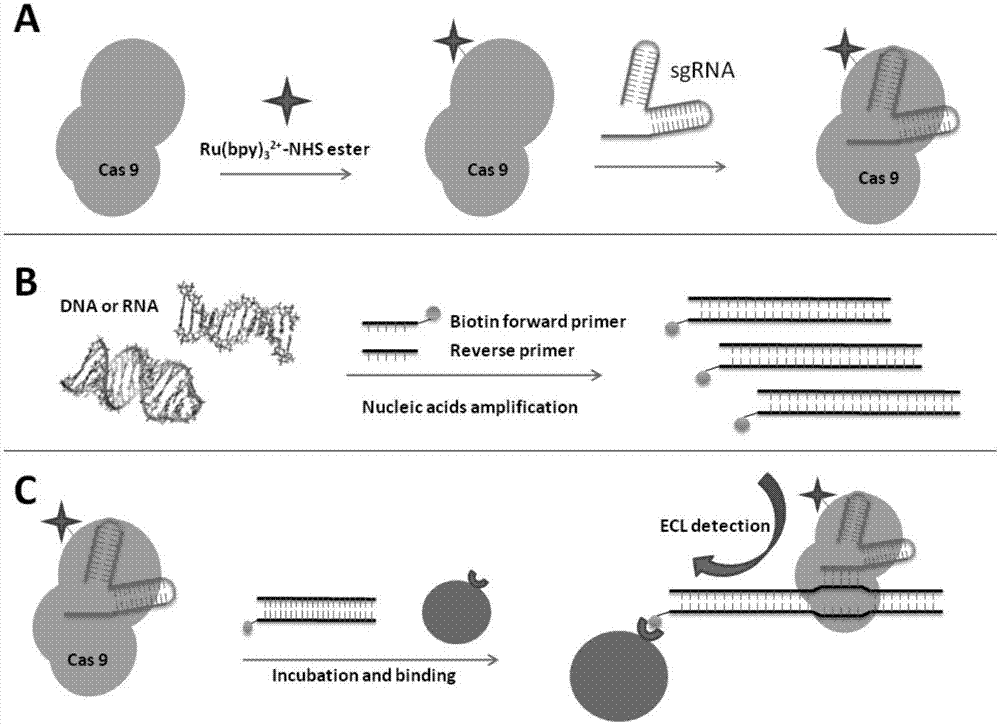
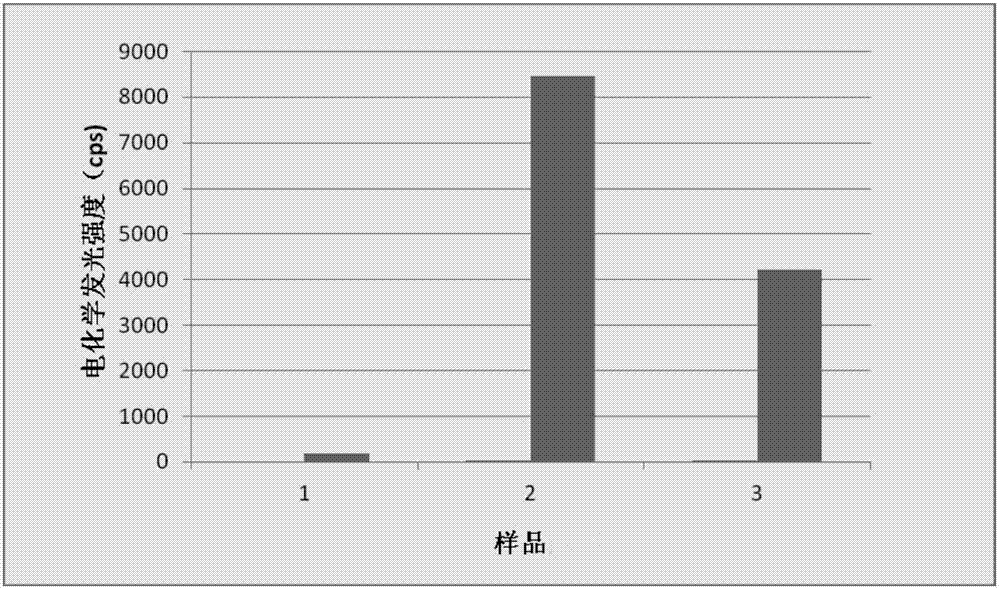
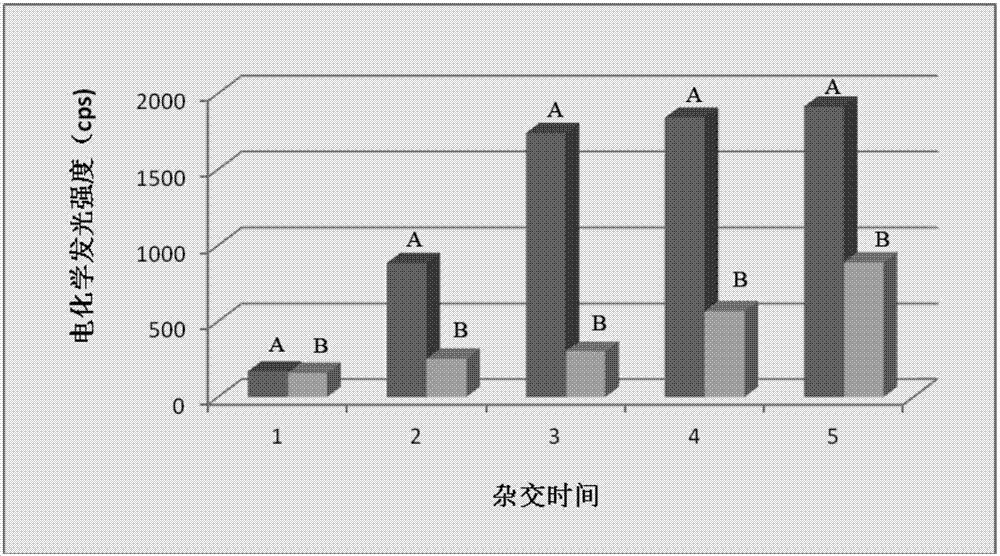
Gene detection probe and gene detection method
A gene detection and probe technology, applied in the field of biochemical analysis, can solve the problems of complex detection steps, slowness, and low hybridization efficiency, and achieve the effects of fast hybridization speed, simple and convenient operation, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment one: Preparation of terpyridine ruthenium-N-hydroxysuccinimide ester (Ru(bpy) 3 2+ -NHS)
[0027] 1. Synthesis of terpyridine ruthenium hexafluorophosphate (Ru(bpy) 2 (dcbpy)(PF 6 ) 2 )
[0028] 0.2 g cis-dichlorobis(2,2'-bipyridyl)ruthenium(II), 0.15 g 2,2'-bipyridine-4,4'-dicarboxylic acid, 0.2 g sodium bicarbonate, 32 mL methanol and Add 8 mL of deionized water into a three-necked flask, install a condenser, heat to 80°C in a silicone oil bath under magnetic stirring, and heat to reflux for 10 hours. The reaction solution gradually changes from purple-brown to orange-red. After the reaction, adjust the pH value of the reaction solution to 4.4 with concentrated sulfuric acid, and cool down in an ice bath for 2 hours in a dark environment to precipitate excess 2,2'-bipyridine-4,4'-dicarboxylic acid , and then vacuum-filtered, and the filtrate was collected to obtain the carboxylated terpyridine ruthenium filtrate. Add 12.5 mL of sodium hexafluorophosp...
Embodiment 2
[0031] Embodiment 2: Ru(bpy) 3 2+ -NHS modification inactivates Cas9 protein
[0032] Add the inactivated Cas9 protein (dCas9 protein, purchased from Shanghai Tulo Harbor Biotechnology Co., Ltd.) to the weakly alkaline PBS buffer to make the final concentration 7 μM / L; add Ru(bpy) 3 2+ -NHS is added in DMF solvent, makes its final concentration be 2mmol / L; Then Ru(bpy) 3 2+ -NHS solution was added to the dCas9 protein solution, and ensured that Ru(bpy) 3 2+ -The molar ratio of NHS to dCas9 protein is 20:1. After slightly shaking and reacting for 1 hour under sealed and dark conditions, add 20 μL of glycine with a concentration of 2 mol / L and incubate for 15 minutes to terminate the reaction. Then equilibrate the reaction system with PBS buffer, and finally filter with Zeba centrifugal desalting column (40kDaMWCO) to remove unreacted Ru(bpy) 3 2+ -NHS. Collect the product Ru(bpy) 3 2+ -NHS-labeled dCas9 protein (dCas9-Ru(bpy) 3 2+ complex), then measure Ru(bpy) 3 ...
Embodiment 3
[0034] Example 3: Preparation of dCas9-Ru(bpy) 3 2+ / sgRNA probe
[0035] 1. Transcription of sgRNA
[0036] The present invention designs a T7 transcription system to transcribe sgRNA. The sgRNA sequence can be divided into two parts according to its function: one part is used to bind to the dCas9 protein to maintain the stability of the dCas9 / sgRNA system; the other part is the targeting region, which is used for gene search and targeting. According to the sequence of the target gene, design the downstream region of the T7 promoter so that the final transcribed RNA sequence contains the complete sequence of the sgRNA.
[0037] According to the fluorescent protein GFP gene sequence, design a pair of specific primers, as follows:
[0038] Forward primer: gaaattaatacgactcactatagggaaggaggacggcaacatcctgttttagagctagaaatagc; (the part in italics is the T7 promoter sequence, the underlined part is the GFP targeting sequence of sgRNA)
[0039] Reverse primer: aaaagcaccgactcggtgc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com