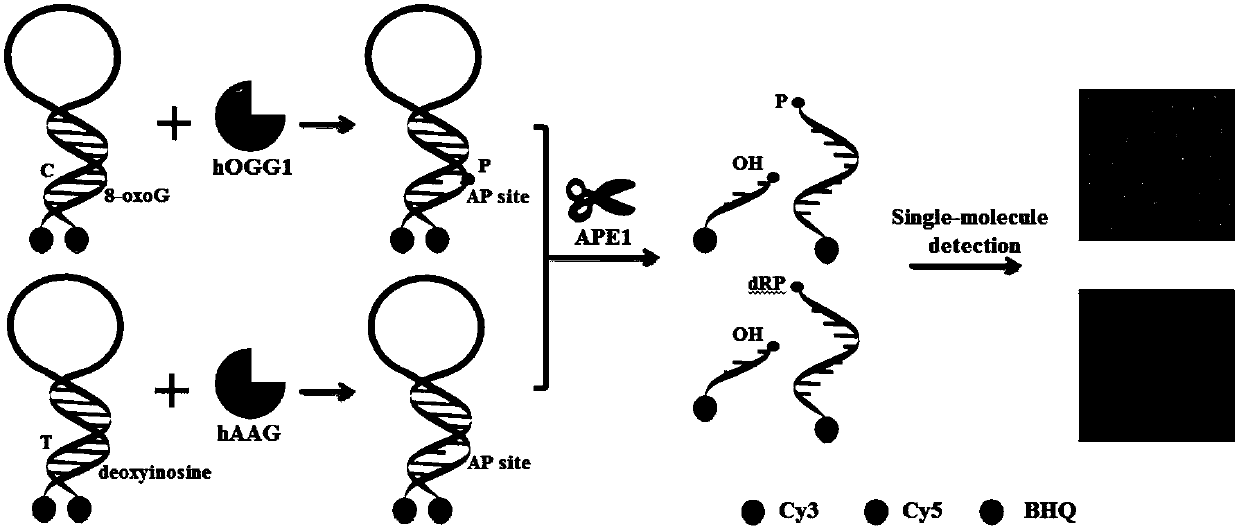
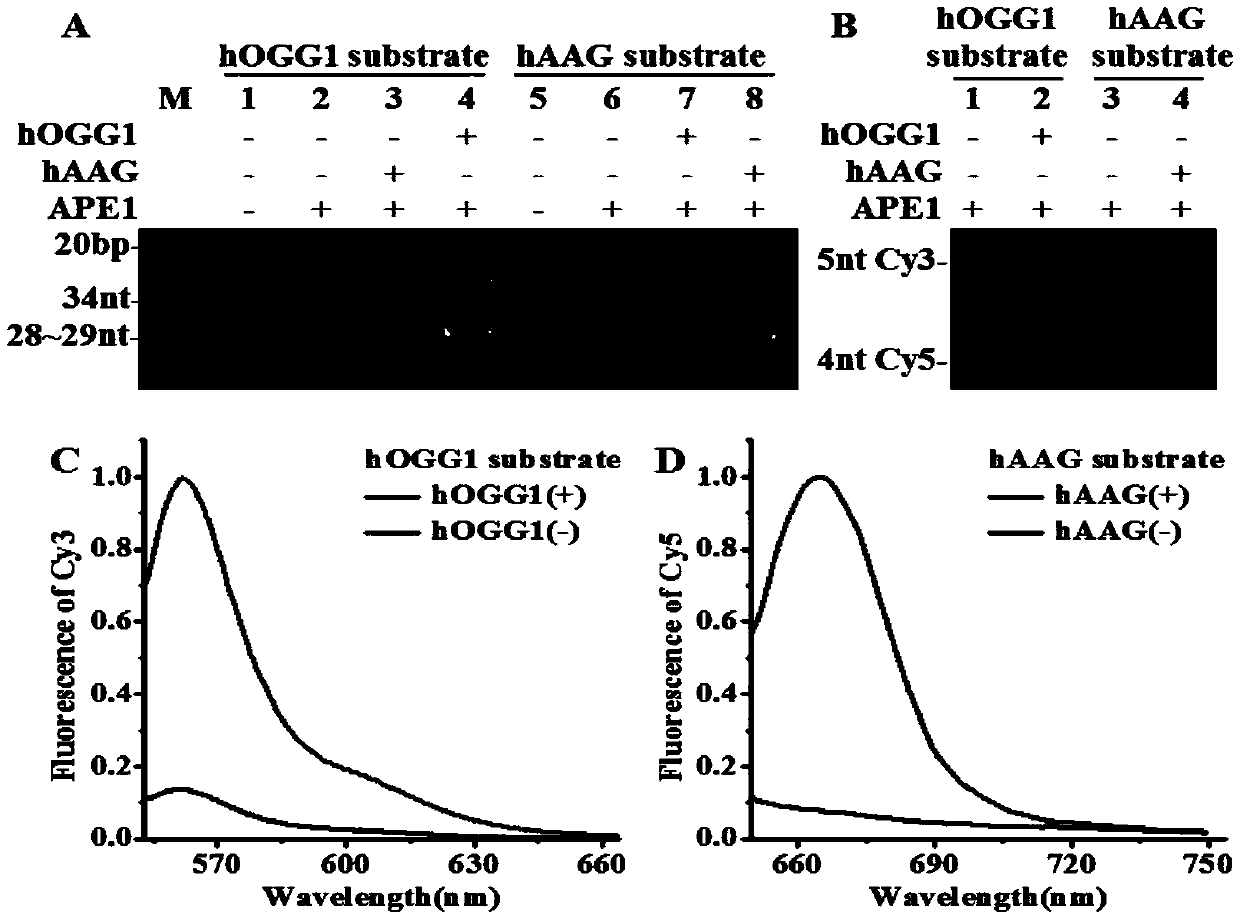
Fluorescence chemical sensor and method for simultaneously detecting diversified DNA (deoxyribonucleic acid) glycosylases on single-molecular levels and application of fluorescence chemical sensor
A chemical sensor and detection method technology, applied in the field of biological analysis, can solve the problems of increased detection complexity and cost, high requirements for the reaction system, and generation of false positive signals, etc., to prevent non-specific reactions, simple preparation and detection methods, The effect of reducing inspection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0062] Experimental method steps
[0063] 1. Molecular beacon preparation: 6 micromoles per liter of hOGG1 probe, 6 micromoles per liter of hAAG probe or 6 micromoles per liter of hOGG1 and 6 micromoles per liter of hAAG probe were added containing 50 millimoles per liter of chloride Sodium, 10 mmol per L Tris-HCl, 10 mmol MgCl, 1 mmol dithiothreitol, pH 7.9 buffer, heat at 95 °C for 5 min, then slowly cool to room temperature, and place the product on ice spare.
[0064] 2. Enzyme digestion reaction: 20 microliters containing 50 mmol per liter of sodium chloride, 50 mmol per liter of potassium acetate, 20 mmol per liter of Tris-Ac, 10 mmol of per liter of magnesium acetate, 10 mmol of potassium chloride, 10 mmol ammonium sulfate, 2 mmol magnesium sulfate, 30 mmol per liter Tris-HCl, 10 mmol magnesium chloride, 1 mmol dithiothreitol, and 0.1% Triton X-100 were added with different concentrations of hOGG1 and hAAG, 0.1 units per microliter of human apurinic / apyrimidinic endon...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com