Method for increasing expression amount of pichia pastoris for secreting and expressing plectasin
A technology of mycelia and Pichia pastoris, which is applied in the field of biotechnology and genetic engineering, can solve the problems that mycelia engineering bacteria cannot fully exert protein secretion expression, retention, and protein folding is too late
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
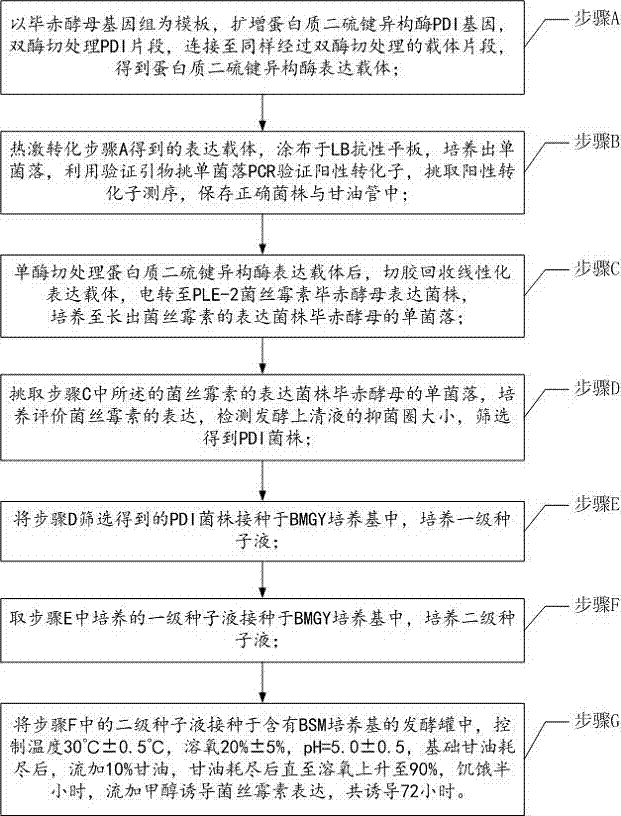
[0062] Example 1 Construction of protein disulfide bond isomerase expression vector
[0063] According to Pichia pastoris protein disulfide bond isomerase PDI gene sequence, design amplification primers
[0064] PDI-F: 5'-TATTCGAAATGCAATTCAACTGGAATAT-3' and
[0065] PDI-R: 5'-GTGAATTCTTAAAGCTCGTCGTGAGCGT-3';
[0066] Add the BstBI restriction site TTCGAA at the 5' end of the gene, and add the EcoRI restriction site GAATTC at the 3' end of the gene;
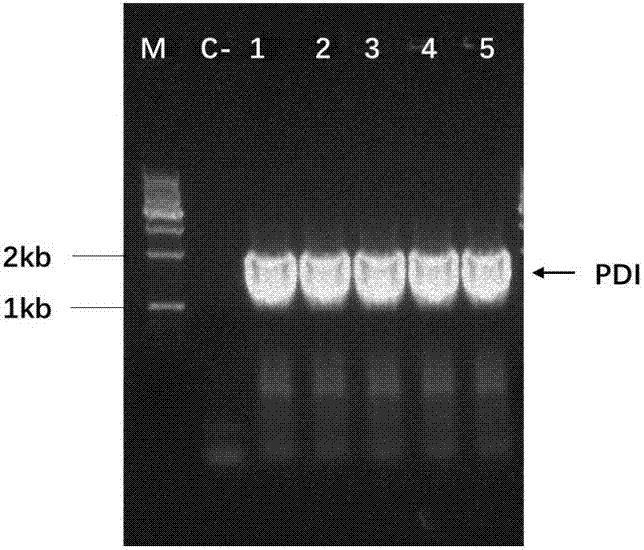
[0067] Using the Pichia pastoris genome as a template, the protein disulfide bond isomerase gene PDI was amplified, and the gene size was 1554bp. The PDI fragment was digested with BstBI and EcoRI, and connected to the pPIC6αA vector fragment that was also digested with BstBI and EcoRI to obtain the protein disulfide bond isomerase expression vector pPIC-PDI. The ligation product was heat-shocked at 42°C and transformed into Escherichia coli Top10 competent cells, spread on an LB resistance plate, and cultured overnight at 37°C...
Embodiment 2
[0071] Example 2 Transformation of Plectasin Engineering Bacteria
[0072] Treat the pPIC-PDI expression vector with PmeI restriction endonuclease, cut the gel to recover the linearized expression vector, electroporate (1.5-2.5kv) to PLE-2 Pichia pastoris expression strain, and spread the bacterial solution on YPD plate (containing 0.1-0.5mg / ml Blasticidin), incubate at 30°C for 2-3 days until a single colony grows.
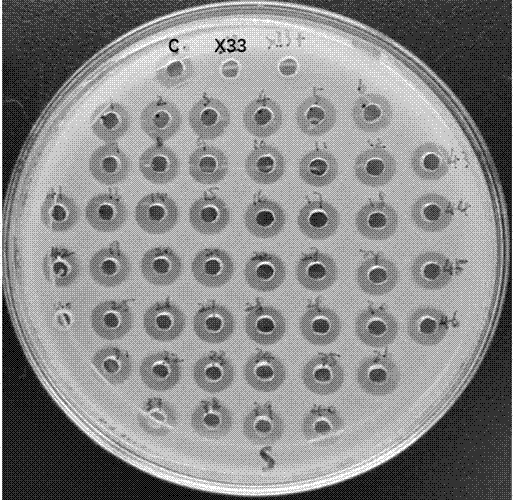
[0073] Pick a single colony, cultivate and evaluate the expression of plectasin, detect the size of the inhibition zone of the fermentation supernatant, and screen to obtain the PDI strain.
[0074] Bacterial inhibition zone detection: Inoculate the indicator bacteria (Staphylococcus aureus CMCC26003) into the MH medium to prepare the indicator bacteria solution. Dilute with normal saline to OD600=2.3, take 100μL diluted indicator bacteria solution to 100mL MH solid medium (temperature 50-55℃), mix well, take 10.5mL solid medium to a standard petri dish, cool an...
Embodiment 3
[0075] Example 3 Induced Expression of Recombinant Plectasin
[0076] A single colony of the PDI strain prepared in Example 2 was picked, inoculated in 25 mL of BMGY medium, and cultured at 30° C. and 220 rpm for 24 hours to prepare a primary seed solution.
[0077] Inoculate 20 mL of primary seed liquid into 200 mL of BMGY medium to prepare secondary seed liquid, and culture at 30°C and 220 rpm for 24 hours.
[0078] All secondary seed liquids were inoculated in a 5L fermenter (2L BSM medium), controlled temperature 30°C±0.5°C, dissolved oxygen 20%±5%, pH=5.0±0.5; after the basic glycerin was exhausted, add 10% glycerol , after glycerol was exhausted until dissolved oxygen rose to 90%, starved for half an hour, fed with methanol to induce the expression of plectasin, induced for a total of 72 hours, centrifuged (6000×g, 5min) to take the fermentation supernatant for detection.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com