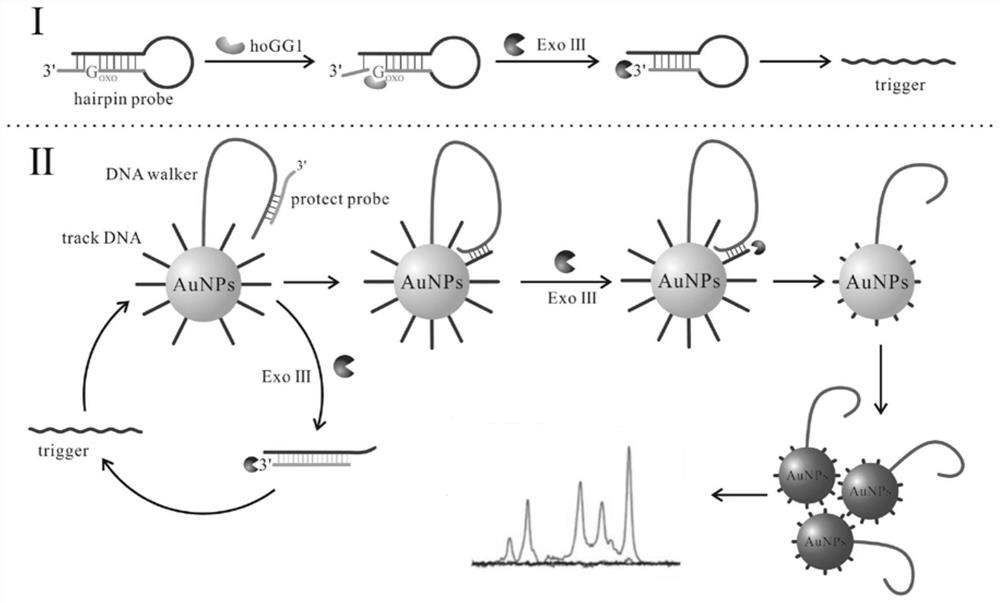
A biosensor for detecting hogg1 activity and its application
A biosensor, active technology, applied in biochemical equipment and methods, microbial determination/inspection, instruments, etc., can solve problems such as low sensitivity and specificity, and achieve high detection sensitivity, mild reaction conditions, and improved reaction speed. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Preparation of gold nanometers.
[0037] (1) Add 200ml of ultrapure water into the three-neck flask;
[0038] (2) Take 500uL of HAuCl with a concentration of 0.04g / mL 4 In the centrifuge tube, add 200ml ultrapure water, stir and heat to boil, stirring speed 450rpm;
[0039] (3) Under stirring conditions, take 3ml of 1% trisodium citrate solution and quickly add it to the solution in step (2). The color of the solution changes from light yellow to wine red. After continuing to heat for 15 minutes, remove the heat source and Slowly cool to room temperature, and store at 4°C for later use.
[0040] According to the absorbance value at 530 nm, the concentration of gold nanoparticles in the above solution is about 0.3 nM.
Embodiment 2
[0041] Example 2 Modification of gold nanometers.
[0042] (1) Walker DNA and Protect DNA were denatured at 90°C, annealed and hybridized at 55°C to form hybrid strands, and stored at 4°C for later use;
[0043] (2) Take 1 mL of the gold nano solution prepared in Example 1 in a centrifuge tube, centrifuge at a speed of 13000 rpm for 10 min. Centrifuge until the supernatant is colorless and transparent, remove the supernatant, and add 300 μL of sterilized water to make the concentration of the nano-gold solution to 3 nM. Transfer it to a 1 mL glass bottle, seal it with tinfoil, and then add 12 μL of DTNB solution (final concentration: 2 μM) while stirring.
[0044] (3) After standing at room temperature for 30 min, add 150 μL of Walker DNA and Track DNA at a concentration of 30 μM, mix well, and place at 4 °C for 24 h.
[0045] (4) Slowly add 50 μL of PBS buffer several times, and add magnetic particles (ddH after soaking in aqua regia 24 hours in advance 2 O washed to neutr...
Embodiment 3
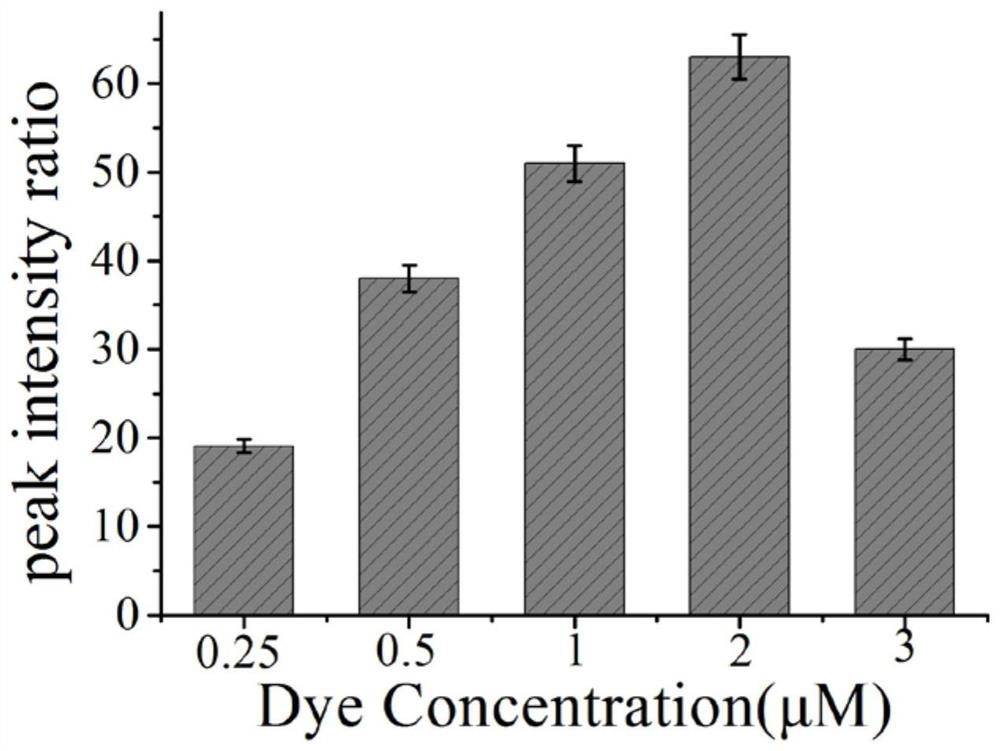
[0048] Example 3 Effects of different concentrations of Raman dyes on hOGG1 glycosidase.
[0049](1) Add 2 μL of ExoⅢ (30U / mL) and 2 μL of hairpin probe (1U / mL) to 6 centrifuge tubes respectively, and add 2 μL of hOGG1 solution (1600U / ml) to 5 test tubes respectively, and 1 test tube Add an equal amount of PBS solution; shake the 6 test tubes for 30 seconds, and put them in a water bath at 37°C for 30 minutes;
[0050] (2) The above reaction solution was added to 20 μL of the surface-modified gold nano-solutions S2-S6 (1 nM) in Example 2, and placed in a water bath at 37°C for 120 min.
[0051] (3) Use a Raman spectrometer to detect the SERS spectrum of the reaction solution obtained in step (2).
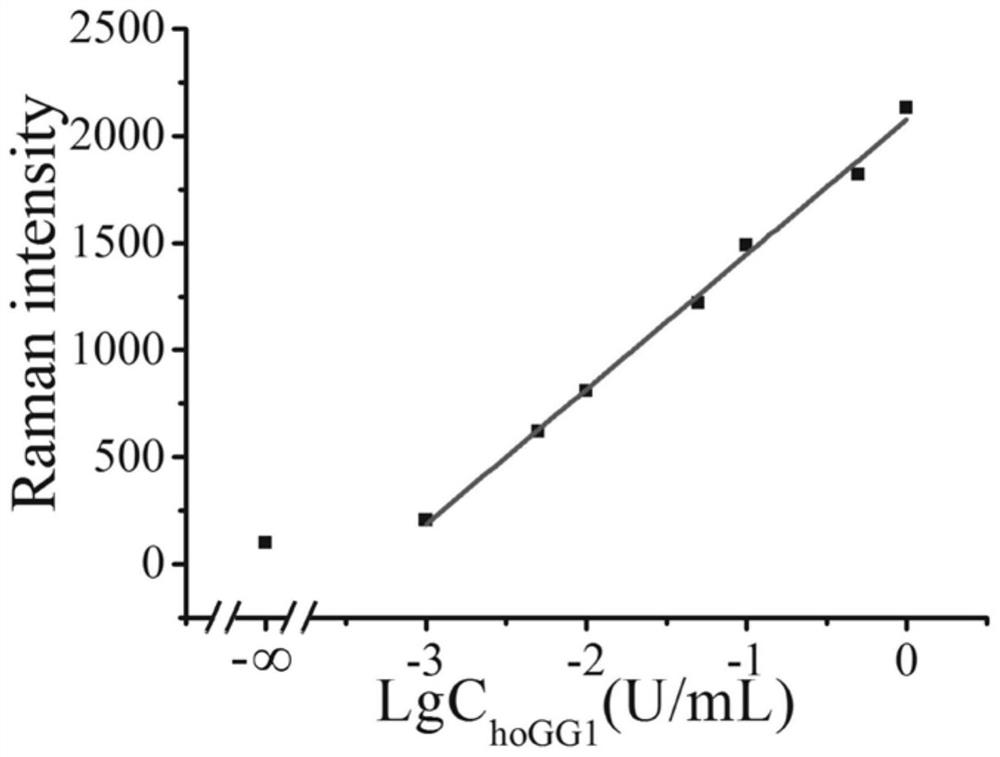
[0052] According to the obtained spectra, the peak area ratios of the characteristic peaks of the SERS spectra of the positive samples and the blank samples were calculated respectively, and the signal-to-background ratios corresponding to the gold nanoparticle probes with differen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com