Enrofloxacin-mimic antigen epitope peptide, and preparation method and application thereof
A technology of antigenic epitopes and mimetic epitopes, which is applied in the direction of analyzing materials through chemical reactions, observing the influence of chemical indicators for material analysis, and material inspection products, etc., which can solve the immunological properties of antigenic epitope peptides Adaptability matching, difficult to obtain detection results, unclear preparation methods and other problems, to achieve high sensitivity, reduce harm to human health and ecological environment, and highlight the effect of application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1, Panning and Identification of ENR Antigen Mimic Epitopes
[0039] (1) Affinity panning of ENR antigen mimotope: the specific method is: use 0.1M NaHCO 3 (pH 8.6) Dilute the anti-ENR monoclonal antibody, and add it to a 96-well enzyme-labeled single well at a final concentration of 80 μg / ml, and coat overnight at 4°C. The next day, after 6 quick washes with TBST [50mM Tris-HCl (pH7.5), 150mM NaCl, 0.1% Tween-20 (v / v)], 350μl of 1% OVA blocking solution was added and blocked at 4°C for 2h. Discard the blocking solution, wash 6 times with TBST, add 120 μl phage peptide library (phage display heptapeptide library, purchased from NEB Company, dilute the phage with TBST, and add 2.0×10 11 pfu), react at 25°C for 50min. The phage in the wells were discarded, quickly washed 10 times with TBST, patted dry, and eluted with 0.2M Glycine-HCl (pH 2.2) for 8 min, and then neutralized with 15 μl of 1M Tris-HCl (pH 9.1). Take 5 μl of the eluted phage to measure the titer,...
Embodiment 2
[0042] Example 2, Sequencing of ENR Antigen Mimotope Encoding Gene and Determination of its Amino Acid Sequence
[0043] The phage identified as ENR antigen mimic epitope by indirect competitive enzyme-linked immunosorbent assay was amplified, and the DNA of the phage was extracted. The operation process is as follows: amplify the target phage, centrifuge the amplified product, transfer 500 μl phage supernatant to a new centrifuge tube; add 200 μl PEG / NaCl to precipitate the phage, let stand for 20 minutes, and centrifuge at 14000 rpm for 10 minutes. Discard the supernatant, resuspend the pellet in 100 μl iodide buffer (10mM Tris-HCl (pH 8.0), 1mM EDTA, 4M NaI), add 250μl absolute ethanol to precipitate, let stand for 15min, and centrifuge at 14000rpm for 10min. After the supernatant was discarded, the precipitate (DNA sequencing template) was washed with 70% ethanol, centrifuged at 14000 rpm for 10 min, the supernatant was discarded, and briefly dried in vacuum. Resuspend th...
Embodiment 3
[0044] Example 3, Application of ENR Antigen Mimotope as Competing Antigen in Enzyme-Linked Immunoassay Method
[0045] (1) Sample extraction
[0046] Remove the fat and connective tissue of the pork, crush it with a mixer, weigh 2 g in a 15 ml centrifuge tube, add 4 ml of 50% methanol-PBS solution, shake on a shaker for 3 min; centrifuge at 4000 rpm for 10 min. Repeat the above shaking and centrifugation steps for the precipitate, combine the supernatant twice, centrifuge again at 10,000 rpm at 4°C for 10 minutes to collect the supernatant, filter it with a microporous membrane (pore size 0.22 μm), dilute the filtrate with PBS to an equal volume, and mix well spare. Take 50 μl of the filtrate and add the sample directly into the microwell for enzyme-linked immunoassay assay.
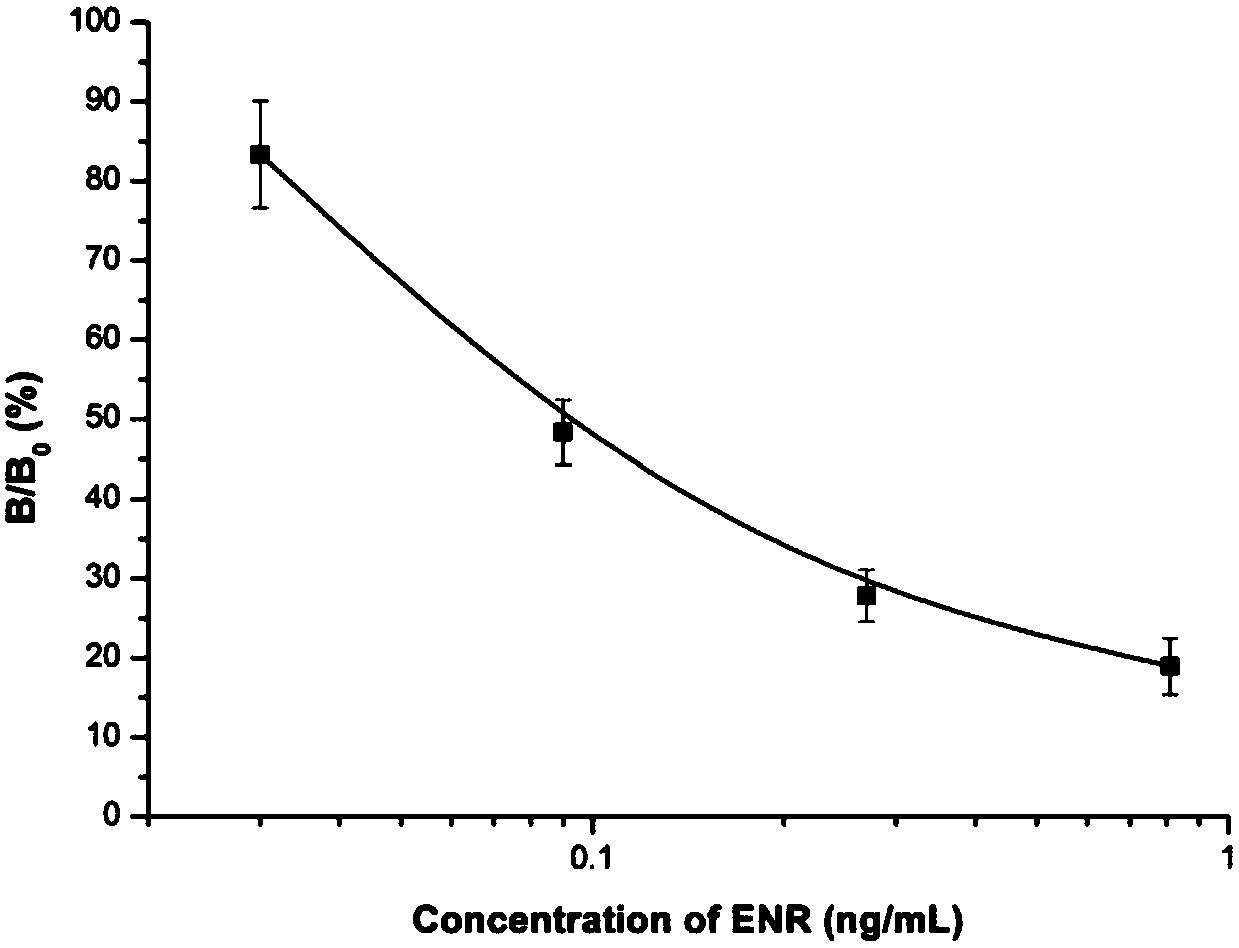
[0048] with 0.1M NaHCO 3 (pH 8.6) Dilute the anti-ENR monoclonal antibody, 1μg / ml coat the microtiter plate, and incubate overnight at 4°C. The next day, wash with PBS...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com