CYP82E10 gene missense mutant M594 capable of reducing nicotine conversion rate as well as application thereof
A CYP82E10, nicotine conversion rate technology, applied in the field of genetic engineering, can solve the problem of reducing the nicotine conversion rate of flue-cured tobacco
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Creation of flue-cured tobacco mutant library
[0019] 1. EMS treatment of flue-cured tobacco seeds
[0020] Seeds of flue-cured tobacco (variety: Yunyan 87) were soaked in 50% commercially available bleach solution for 12 minutes, then quickly washed with deionized water, and soaked in deionized water for 12 hours. Discard deionized water and add an equal volume of 0.5% EMS (ethyl methanesulfonate) for 12 hours. Discard the treatment solution, add deionized water to wash 6-8 times, about 1 minute each time. The seeds were collected in a Buchner funnel and drained for later use.
[0021] 2. Field planting of M1 plants
[0022] After EMS treatment, the seeds are sown on floating trays, one seed per hole, and transplanted to the field after emergence, and managed with normal agronomic measures. After budding, single plant was bagged and harvested to obtain M2 seeds.
[0023] 3. Mutant Genomic DNA Extraction and Sample Mixing
[0024] Use the kit to extract genomic D...
Embodiment 2
[0028] CYP82E10 Gene Mutant Screening
[0029] 1. Tilling primers
[0030] CYP82E10 The gene has two exons, and the mutants in the first exon region were screened by Tilling technique. According to the genome sequence of the target gene,
[0031] The forward primer is CYP82E10 _Tilling_F:GTCAAATACCACCTCTTAATAGTAA,
[0032] The reverse primer is CYP82E10 _Tilling_R: AAAAGTCCCTATTGGTAGGAAGTGC.
[0033] 2. PCR amplification conditions
[0034] The PCR reaction system is as follows: the total volume is 10 μL, including 1.0 μL of 20 ng / μL DNA sample, 1.0 μL of 10×PCR buffer, 0.8 μL of dNTPs, 0.16 μL of each primer, 0.1 μL of Taq DNase, and 6.78 μL of ddH2O.
[0035] The PCR reaction program is as follows: pre-denaturation at 95°C for 3 minutes; denaturation at 94°C for 30 seconds; Anneal at ℃ for 30 seconds, extend at 72℃ for 90 seconds, run 40 cycles; finally extend at 72℃ for 5 minutes. PCR amplification products can be stored at 4°C.
Embodiment 3
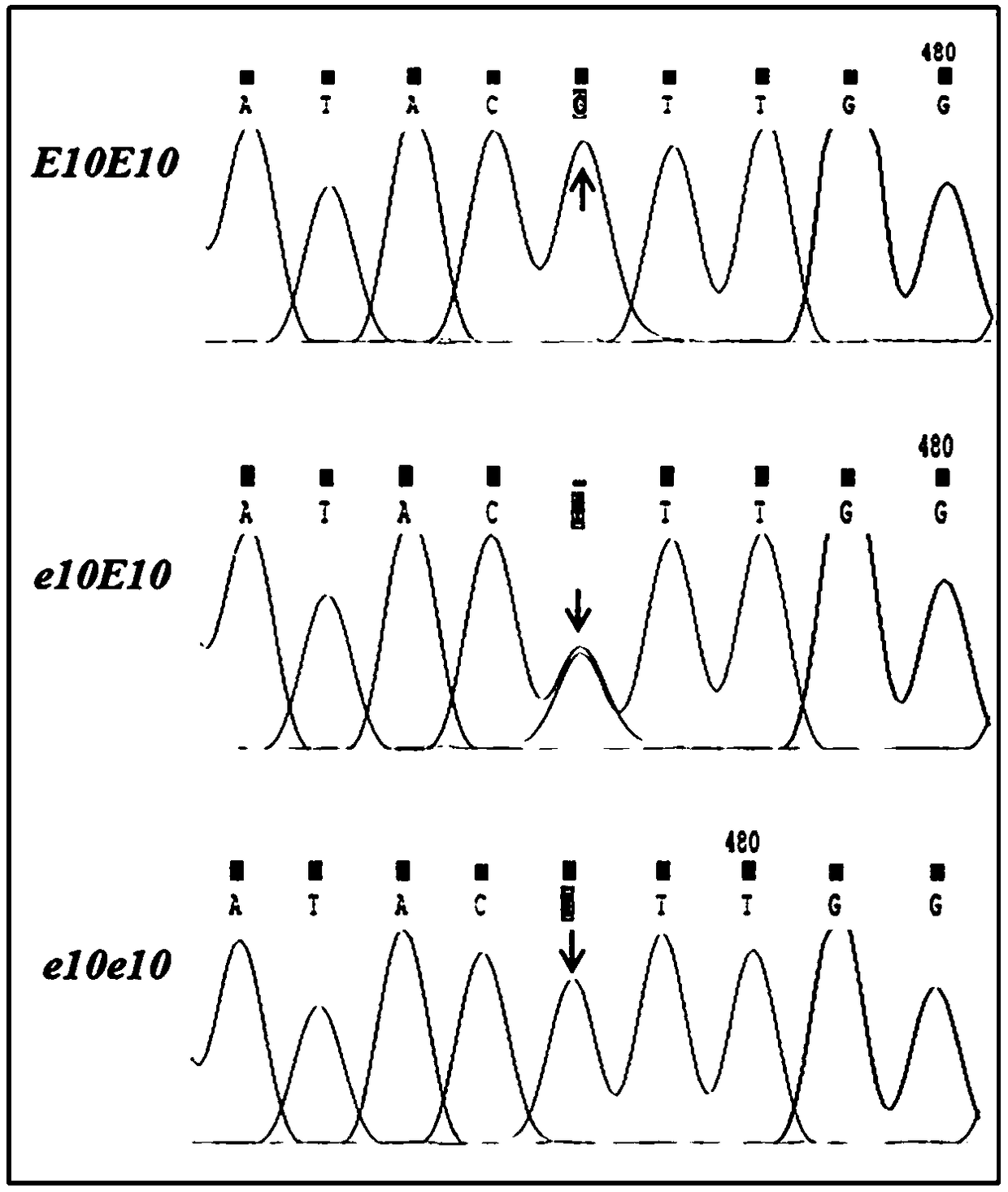
[0041] CYP82E10 Gene missense mutation verification
[0042] 1. Genomic DNA extraction and PCR amplification of M3 generation mutants
[0043] According to the Tilling screening results of the M2 generation plants, select CYP82E10 Seeds (M3 generation) of a single plant with mutations in the gene target region were sown in seedling trays. The genomic DNA of seedling leaves was extracted by kit method. by CYP82E10 _Tilling_F and CYP82E10 _Tilling_R primer, amplified with genomic DNA as a template CYP82E10 The first exon region of the gene. The PCR reaction system is as follows: the total volume is 25 μL, including 1.0 μL of 20 ng / μL DNA sample, 2.5 μL of 10×PCR buffer, 2 μL of dNTPs, 0.5 μL of each primer, 0.3 μL of Taq DNase, ddH 2 O 18.2 μL. The PCR reaction program is as follows: pre-denaturation at 95°C for 3 minutes; denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds (1°C drop per cycle), extension at 72°C for 90 seconds, and 30 cycles; exte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com