DNA library kit for sequencing and construction method of DNA library for sequencing
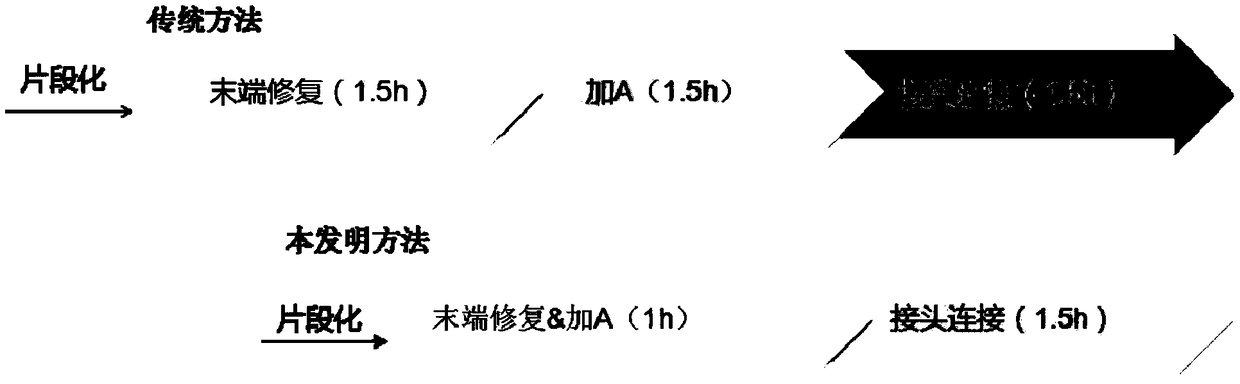
A library construction and kit technology, applied in DNA/RNA fragments, recombinant DNA technology, libraries, etc., can solve the problems of time-consuming, high consumption of purification reagents, and high cost of the overall library construction process, and achieve high ligation efficiency and effective library. High yield and control of nucleic acid contamination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
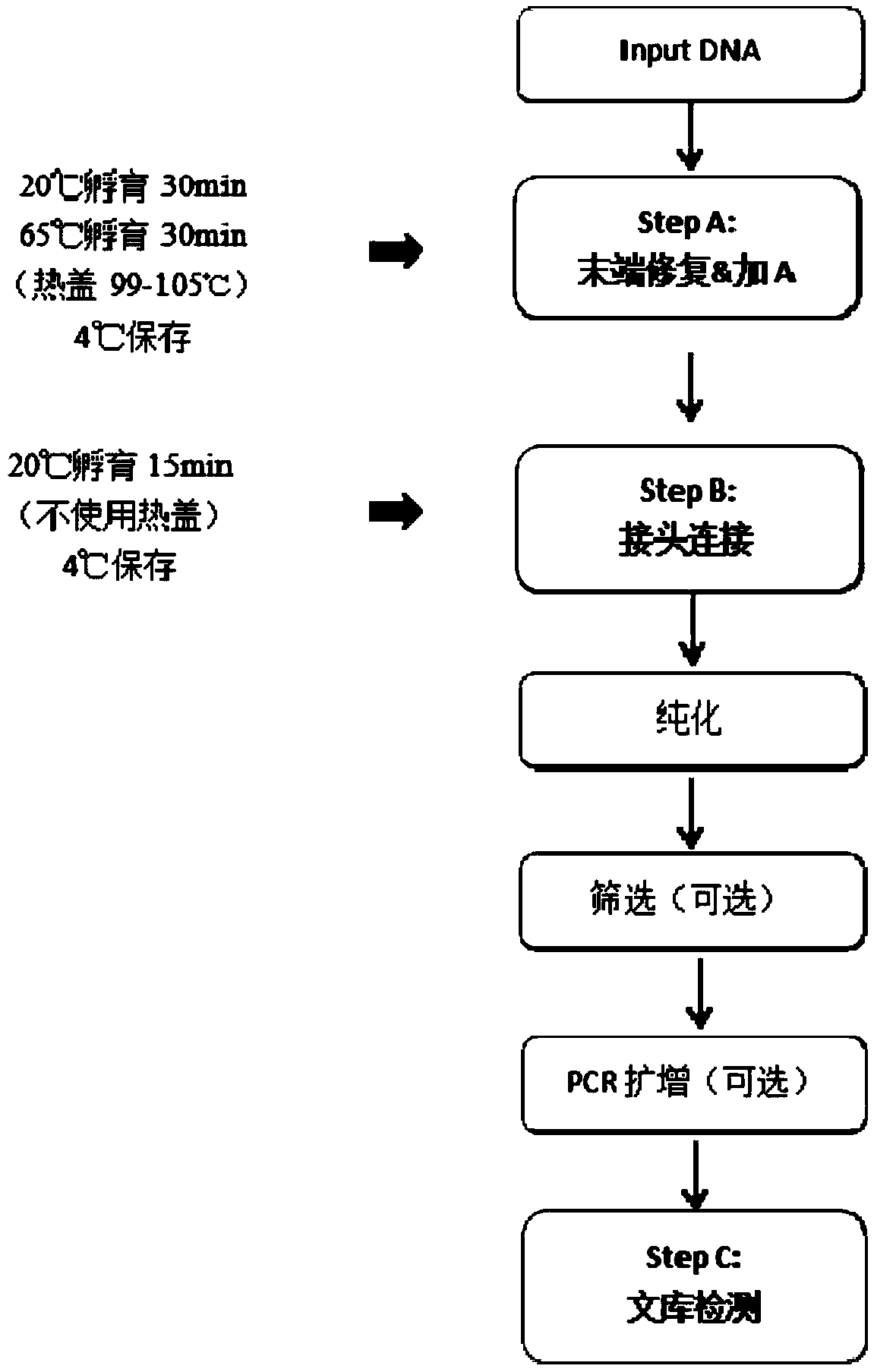
[0092] The library construction method based on the Illumina sequencing platform, the steps are as follows:
[0093] (1) Sample preparation: Escherichia coli (E.coli) genome was extracted from E. coli LB medium, and the extraction method was Omega whole genome extraction kit. The extracted large fragments of DNA need to be physically interrupted by Covaris, and the interrupted DNA sequence is a small fragment of DNA of about 200bp. The fragmented DNA fragments were purified by Ampure XP magnetic beads, diluted to a concentration of 10 ng / μl with TE buffer, and OD260 / OD280=2.0.
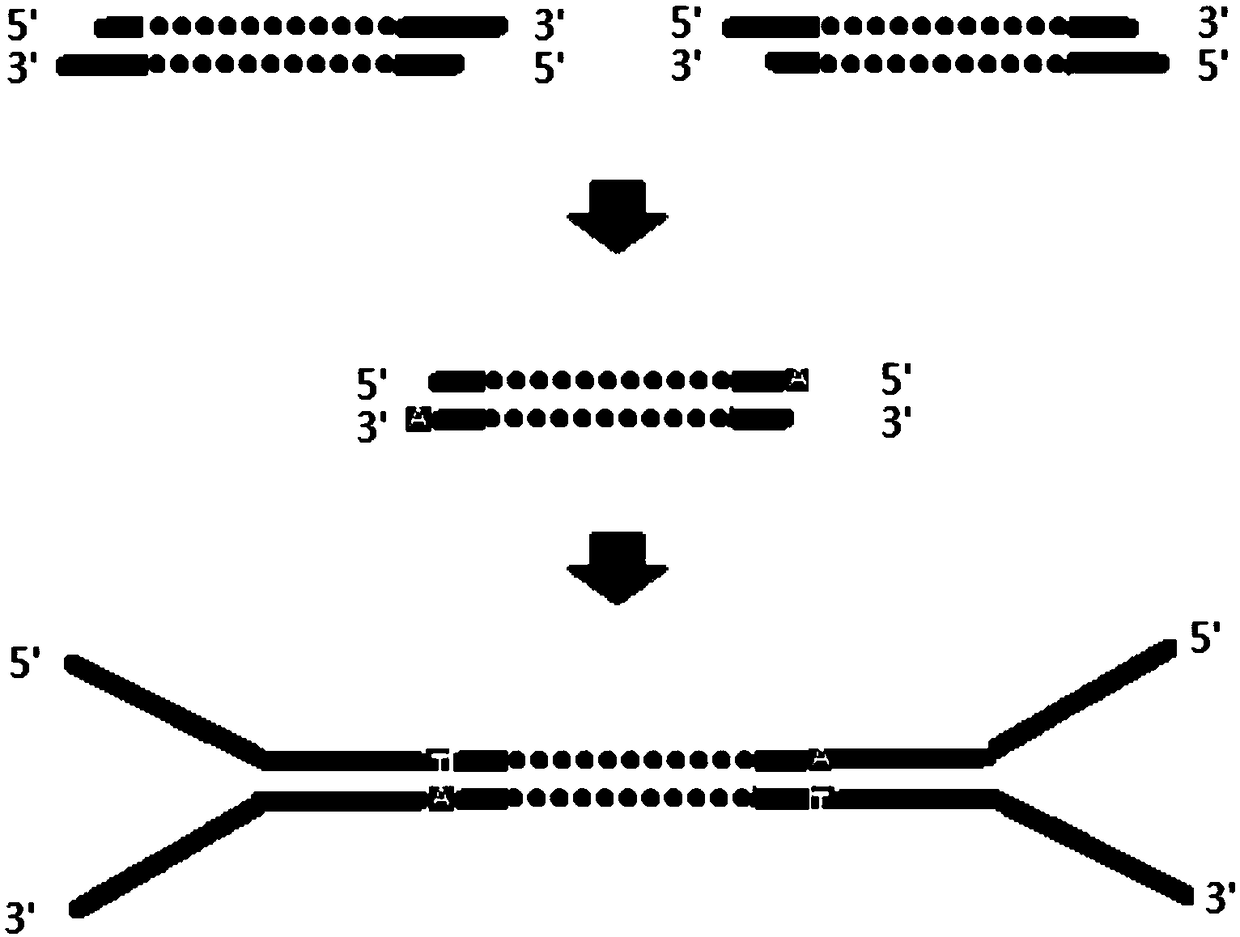
[0094] (2) Fragmented DNA (DNA Fragment) end repair and A: Dilute the fragmented DNA interrupted by step (1) to an appropriate concentration, use end repair and add A reagents for end repair and insert into the fragmented DNA An adenosine (A base) is added to the 3' end of the sequence.
[0095] The reaction system is shown in Table 1.
[0096] Table 1 reaction system
[0097]
[0098] Among the...
Embodiment 2
[0136] The difference from Example 1 is that the end repair and the components of A buffer: Tris-HCl 10mM, ATP 1mM, MgCl 2 10mM, dNTP 100μM, dATP 1mM;
[0137] End repair and A enzyme use 5U of Bst DNA polymerase;
[0138] End repair with plus A: incubate at 20°C for 30 minutes, and at 37°C for 30 minutes.
[0139] Components of ligation reaction buffer: Tris-HCl 10mM, ATP 1mM, MgCl 2 10mM, 2% PEG4000;
[0140] The ligase is a mixture of equal volumes of 50 U of T4 DNA ligase, 1 U of Bst DNA polymerase, and 1 U of T4 polynucleotide kinase.
[0141] The ligation condition is: incubate at 20°C for 30 minutes.
[0142] The rest are the same as in Example 1, and the effect of building a library is basically the same as in Example 1.
Embodiment 3
[0144] The difference from Example 1 is that the end repair and the components of A buffer: Tris-HCl 50mM, ATP 3mM, MgCl 2 50mM, dNTP 300μM, dATP 5mM;
[0145] End repair and A-adding enzyme use 5U of Klenow enzyme (3'-5'EXO-) and 5U of T4 DNA polymerase in a volume ratio of 1:1 mixed enzyme;
[0146] End-repair with Add A: Incubate at 30°C for 15 minutes and at 65°C for 15 minutes.
[0147] Components of ligation reaction buffer: Tris-HCl 50mM, ATP 3mM, MgCl 2 50mM, 8% PEG6000;
[0148] The ligase is T4 DNA ligase with 200 U of ligase.
[0149] The ligation conditions were: incubate at 37°C for 15 minutes.
[0150] The rest are the same as in Example 1, and the effect of building a library is basically the same as in Example 1.
[0151] In addition, in the present invention, the length of the linker sequence can be other commercially available linkers suitable for the Illumina platform, or it can be the 20-50 bp base sequence of the linking part of the linker and the i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com