CRISPR/Cas12a gene editing system and application thereof
A gene editing and gene technology, applied in the field of gene editing systems, can solve the problems of long cycle, high cost and low recombination frequency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
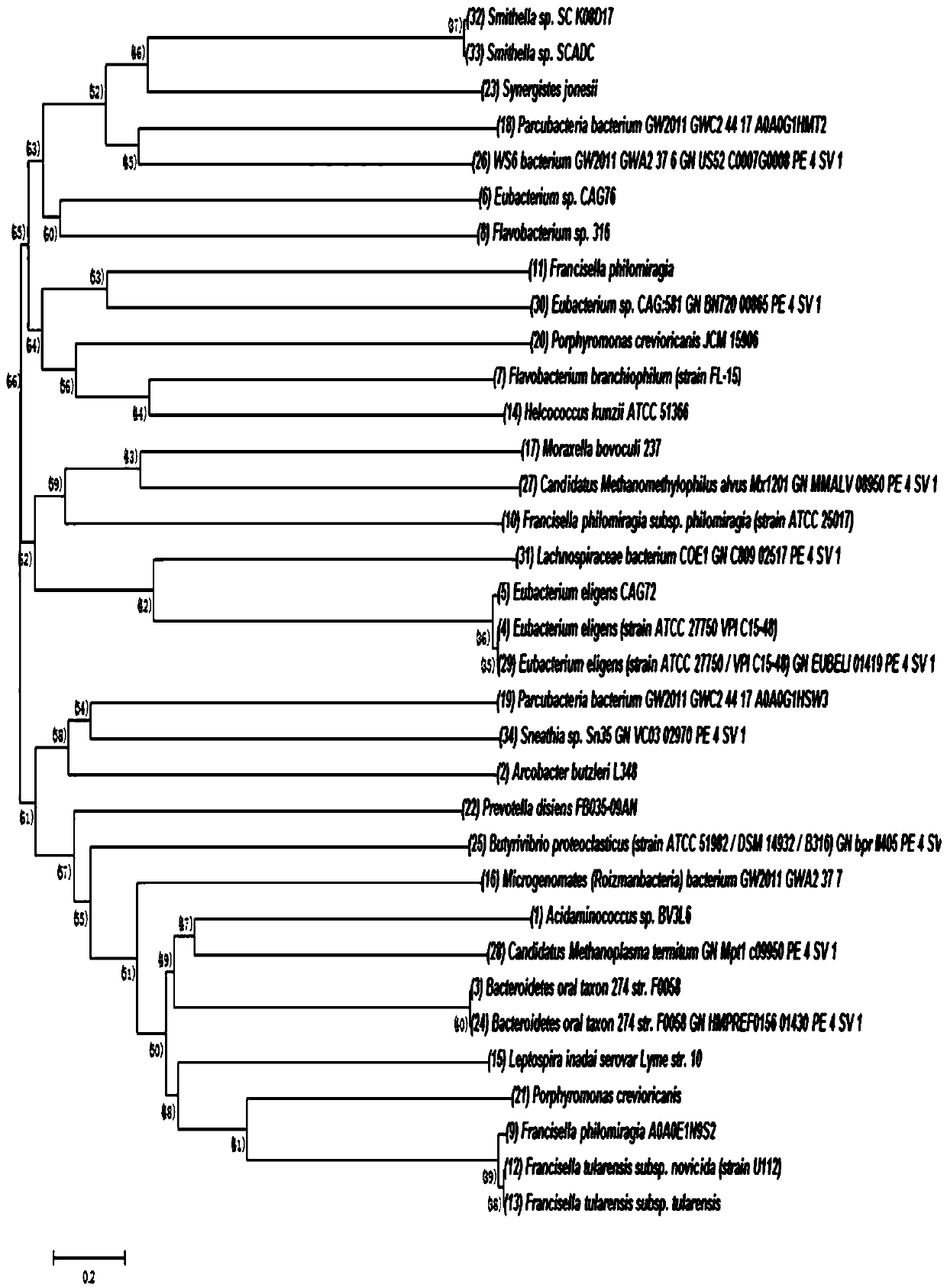
[0036] 40 Cas12a protein sequences were collected from Uniprot, and a phylogenetic tree was constructed for analysis, among which LiCas12a was proved to have double-stranded DNA cutting activity. The specific method is as follows:
[0037] To find new functional Cas12a for genome editing, we collected 40 Cas12a protein sequences from Uniprot (https: / / www.uniprot.org) and analyzed these sequences together with AsCas12a, FnCas12a and LbCas12a by phylogenetic tree ( figure 1 ). Considering the different evolutionary origins and lengths of Cas12a proteins, five Cas12a proteins from Arcobacter butzleri L348, Eubacterium eligens, Francisella philomiragia subsp. philomiragia, Helcococcus kunzii and Leptospira inadaiserovar Lyme were selected as candidates. Among these five candidates, LiCas12a demonstrated DNA cleavage activity.
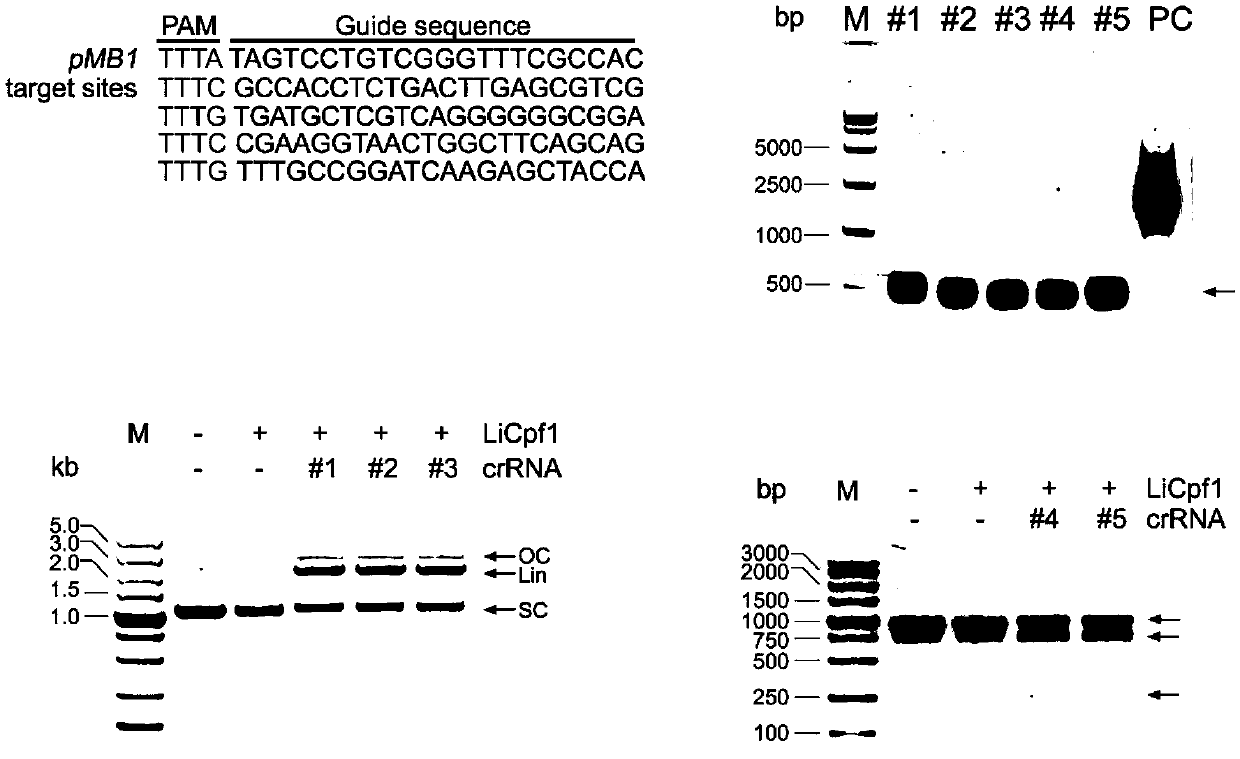
[0038] First clone LiCas12a into pGEX-6p-1 vector. The protein LiCas12a was expressed and purified using Ni affinity chromatography from E. coli BL21....
Embodiment 2
[0041] In order to verify the activity of LiCas12a in mammalian cells, that is, whether it has the function of cutting DNA, we verified it through the following experiments.
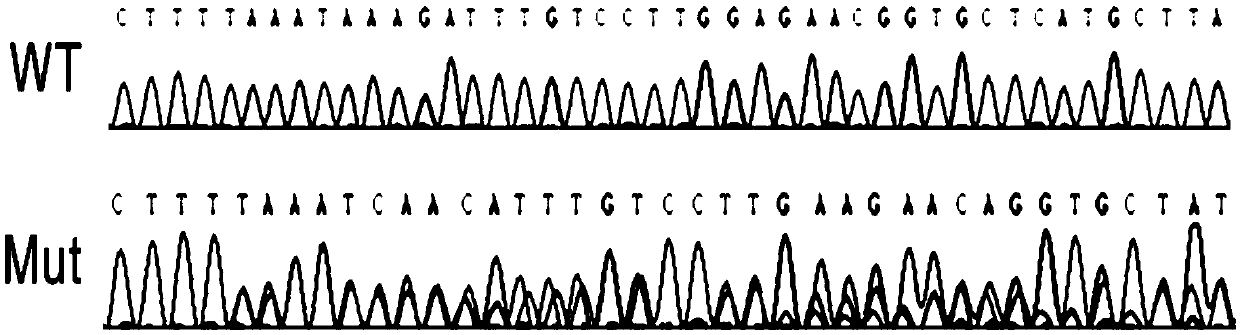
[0042] First, the screened LiCas12a gene (see SEQ ID No: 1 in the sequence listing) was cloned into the plasmid pLiCas12a with crRNA and N23 to select the DNMT1 gene as the knockout target (see the sequence listing in SEQ ID No: 4). The constructed LiCas12a plasmid targeting DNMT1 was transfected into 293T cells, and then the GFP-positive cells were sorted out for monoclonal culture. The genomic DNA of the monoclonal cells was extracted and used as a template to amplify DNMT1, and sequenced for verification.
[0043] The DNMT1 gene was successfully disrupted in 9 out of 10 monoclonal cell lines (90%). image 3 Representative sequencing results are shown. In Western blot analysis, DNMT1 expression was completely disrupted in DNMT1-deficient cells (Mut) compared with wild-type cells (WT) ( Figure 4 ). ...
Embodiment 3
[0045] CRISPR-LiCas12a system-mediated point and back mutations in mammalian cells.
[0046] TERT, encoding telomerase reverse transcriptase, is a component of telomerase, a ribonucleoprotein polymerase that maintains the ends of telomeres by adding the telomeric repeat TTAGGG21. The TERT promoter is frequently altered in many tumor types such as melanoma, hepatocellular carcinoma, glioma, urothelial carcinoma and renal cell carcinoma. The mutations of chr5, 1,295,228C>T and 1,295,250 C>T in the TERT promoter were the most important mutations. We tested the genomic status of TERT in bladder and hepatocellular carcinoma cell lines, selecting T24 and Huh7 as editing targets.
[0047] Two independent crRNAs were designed for the promoter region of the TERT gene (for the sequence, see SEQ ID No: 5 in the sequence listing). The bladder cancer cell line T24 (chr 5:1295228T) and the hepatocellular carcinoma cell line Huh7 (chr 5:1295228C) were selected as the editing objects. The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com