Long-chain non-coding RNA T5120 derived from arabidopsis thaliana and use thereof
An RNAT5120, long-chain non-coding technology, applied to long-chain non-coding RNAT5120 and its field of improving nitrate nitrogen assimilation and utilization, can solve problems such as reports on nitrate nitrogen signal and metabolism research, and improve nitrogen utilization rate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
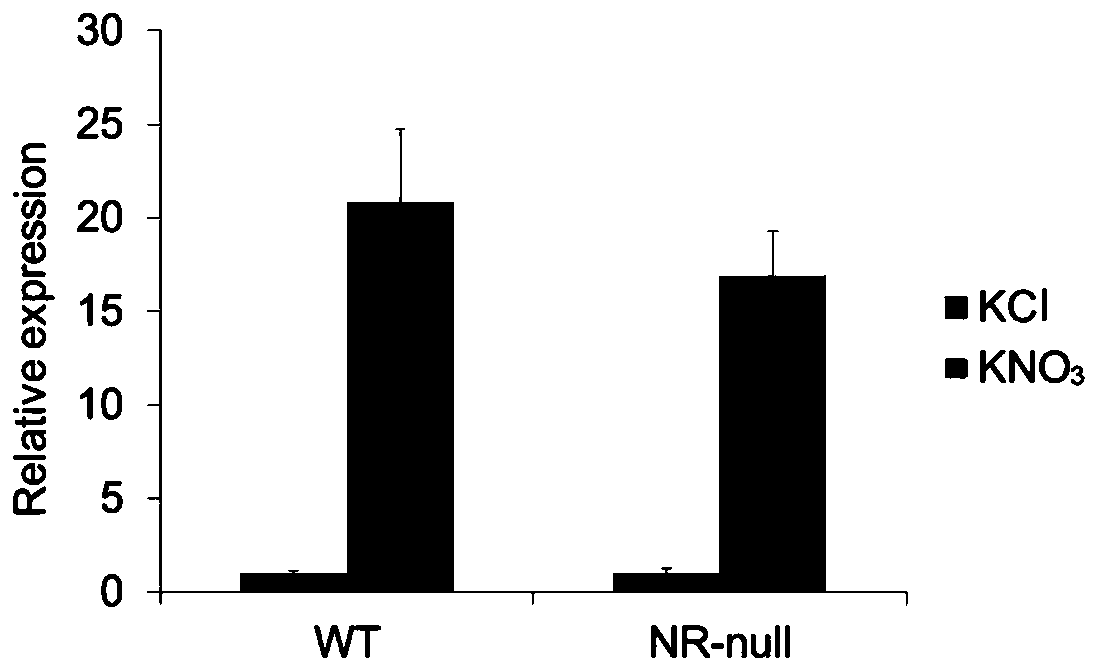
[0050] Example 1: Identification of nitrate nitrogen response lncRNA T5120
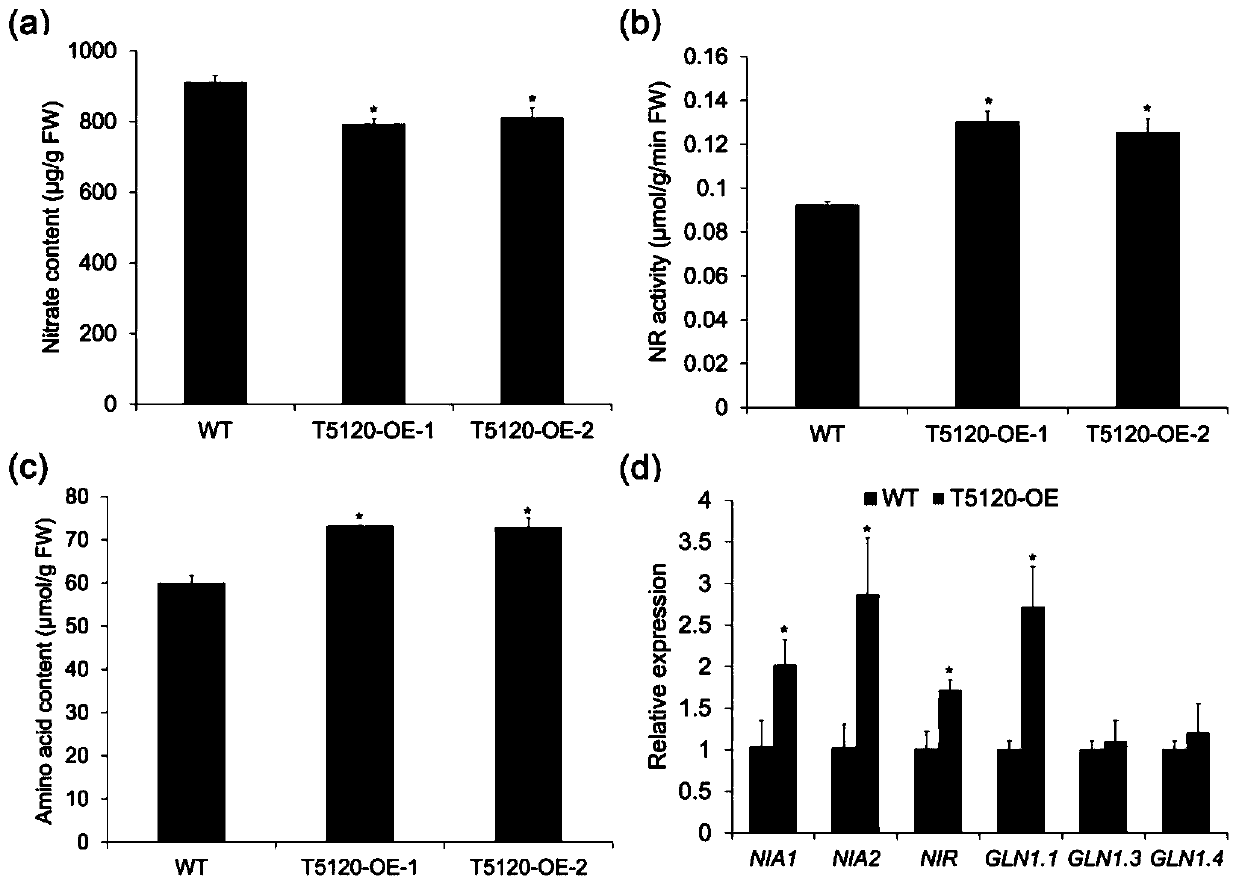
[0051] Arabidopsis wild-type in 2.5mM NH 4 Grow on Suc medium for 7 days with 10 mM KNO 3 Or 10mM KCl (as a control) was treated for 2 hours for RNA-seq sequencing. Analyze the results of high-throughput sequencing to screen lncRNAs that can respond to nitrate nitrogen, and the expression of lncRNA T5120 is affected by NO 3 - The most obvious changes after treatment. In order to verify the results of RNA-seq sequencing, the wild type and NR-null (nia1nia2) mutants were grown under the above culture conditions and treated accordingly, the roots were taken and RNA was extracted, and the RNA was reverse-transcribed into cDNA using a kit. Using cDNA as a template, qPCR experiments were carried out, and it was found that in NO 3 - The expression of T5120 in the wild type and NR-null mutant after treatment was significantly increased, indicating that the expression of T5120 was directly affected by NO...
Embodiment 2
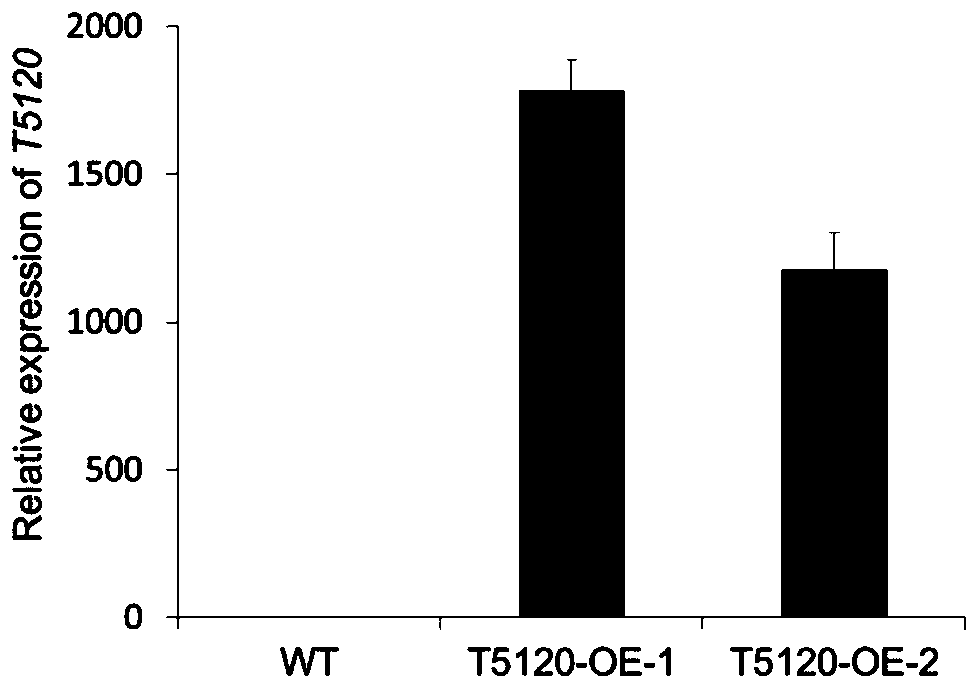
[0058] Example 2: Construction of lncRNA T5120 plant expression vector
[0059] The wild-type Arabidopsis thaliana was grown on 1 / 2 MS medium for 7 days, and the whole seedlings were taken and genomic DNA was extracted. Using the extracted DNA as a template, a high-fidelity DNA polymerase was used to amplify the gene fragment of T5120. The primer sequence used in PCR was: upstream primer: 5'-ACACGGATCCTGACGTCGGCAACATATAGC-3', the nucleotide sequence of which was shown in SEQ ID NO.2 Shown; downstream primer: 5'-ACACGTCGACGGGCGGTATATTGACTCGAA-3', its nucleotide sequence is shown in SEQ ID NO.3. The PCR product was electrophoresed in 1% agarose, and the target band was found according to the DNA Marker and the gel block was excised. Utilize the gel recovery kit (purchased from Omega Company) to recover the target DNA, carry out enzyme digestion (BamHI and SalI) to the recovered target DNA and the plant expression vector pZP211, and then digest the DNA fragment and vector with T...
Embodiment 3
[0065] Embodiment 3: Identification of transgenic positive plants
[0066] Pick the Agrobacterium cells stored at -80°C in 250mL LB culture medium, place them on a shaker at 28°C, and culture them with shaking for 16 hours. Agrobacterium was used to infect Arabidopsis wild-type plants by flower dipping method. After the plants are mature, collect the seeds, and spread the seeds in the K-containing + On the resistant 1 / 2 MS plate, the screened positive seedlings are the transgenic line T1 generation. The selected T1 generation seeds were transplanted into vermiculite for cultivation, and the T2 generation seeds were harvested. Seeds of the T2 generation in the K + The resistant 1 / 2 MS plate was cultured, and its survival rate was found to be 3 / 4. The surviving T2 plants were moved to vermiculite for culture, and the T3 generation seeds were harvested from a single plant. The seeds of the T3 generation were placed in the K + The resistant 1 / 2 MS plates were cultured, and th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com