Biocontrol bacterium for mulberry diseases and application thereof
A technology for mulberry and disease, which is applied in the field of biocontrol bacteria for mulberry bacterial diseases, can solve the problems of soil environment deterioration, pathogenic drug resistance enhancement, difficult control, etc., and achieve the effect of avoiding soil environment deterioration and good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
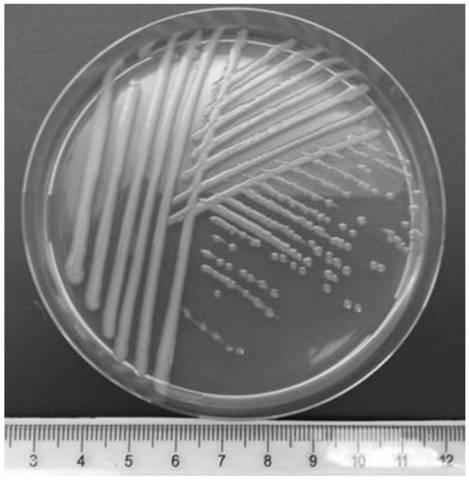
[0042] The separation and purification of embodiment 1 root endophyte
[0043] 1. Experimental method
[0044] 1. Isolation and purification of root endophytes
[0045] Take the freshly collected mulberry root tissue and rinse it with tap water, cut into about 3-4 cm root segments and place them in a funnel with a switch (the funnel has a filter screen), and then add 75 % alcohol, soaked for about 15s, then drained the alcohol, treated with mercuric chloride for 5 minutes in the same way, and then rinsed with sterile water for 3 to 4 times, took out the root segment and put it on the workbench to air dry, then cut the root segment with sterile tweezers. The cortex is torn off to expose the xylem, and then the longitudinal strips of the xylem are scraped off with a sterile scalpel and placed in a sterile mortar. After adding about 5mL of sterile water, grind it thoroughly. Use a pipette to take 1mL of the grinding solution and add 9mL of it. In sterile water, release 5 times ...
Embodiment 2
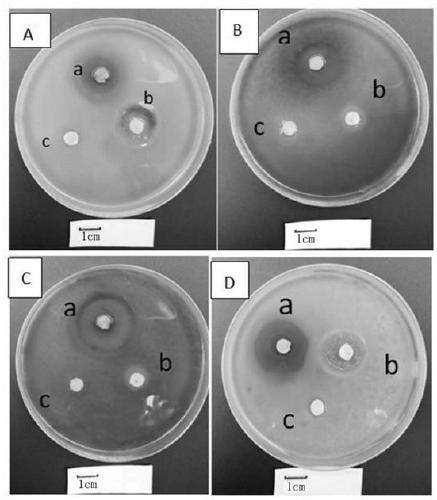
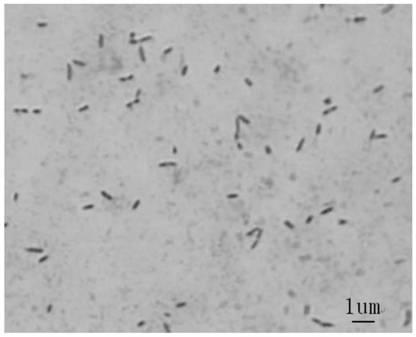
[0057] Identification of embodiment 2 antagonistic bacteria
[0058] The culture characteristics of the colony plate, Gram staining and various physiological and biochemical index tests are important basis for determining the classification status of bacterial strains. Because rRNA gene sequence is highly conserved, it can be used as an important reference standard for evolutionary analysis.
[0059] 1. Experimental method
[0060] The screened antagonistic strain (YD-001) was inoculated in beef extract peptone medium, cultured at 28°C for 2 days, and the colony morphology, growth status, soluble pigment production, etc. were observed and recorded. Gram staining, morphological observation, contact enzymes, aerobicity and other related physical and chemical parameters were determined according to the "Common Bacterial System Identification Manual" and "Berger Bacterial Identification Manual".
[0061] The entire genome of the antagonistic bacteria (YD001) was sequenced, and t...
Embodiment 3
[0070] Pot control of embodiment 3 antagonistic bacteria YD-001
[0071] 1. Experimental method
[0072] The soil used in the pot experiment was humus soil, and the mulberry seed variety was selected as the susceptible variety "Guisang 12". The pathogens of mulberry wilt were isolated and preserved in this experiment, including Enterobacter cloacae, Pantoea, Raueria morulus, and Klebsiella. Mulberry seedlings are 50d seedlings. Experimental group, for testing mulberry seedlings with 10 6 The cfu / mL antagonistic bacteria YD-001 strain suspension soaked the roots for 1 min, and then poured the bacterial suspension once every other week for a total of 2 times. After 20 days, the wounded root method was used to inoculate the roots for 10 days. 8 cfu / mL pathogenic bacteria suspension 10ml. The blank control group was only inoculated with pathogenic bacteria. 30 mulberry seedlings per treatment. After 20 days, investigate and record the incidence of mulberry plants, and calcul...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com