Gene sequencing data simulation system and method for simulating crowd background information
A background information and data simulation technology, applied in the field of data science, can solve the problems of large limitations, inability to simulate population polymorphism, and high sample extraction cost, so as to improve operating efficiency, enrich variation simulation functions, and save computing time. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
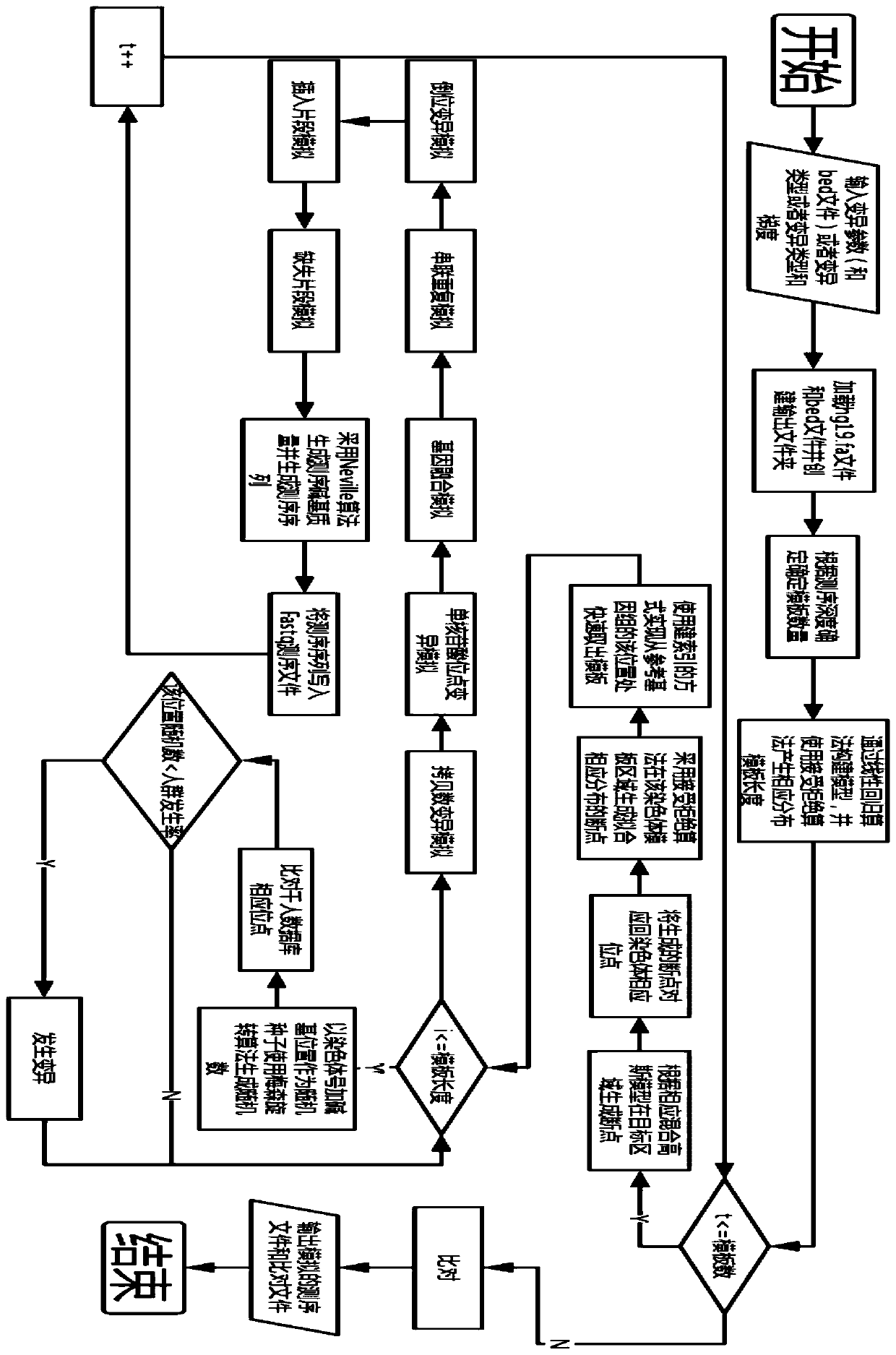
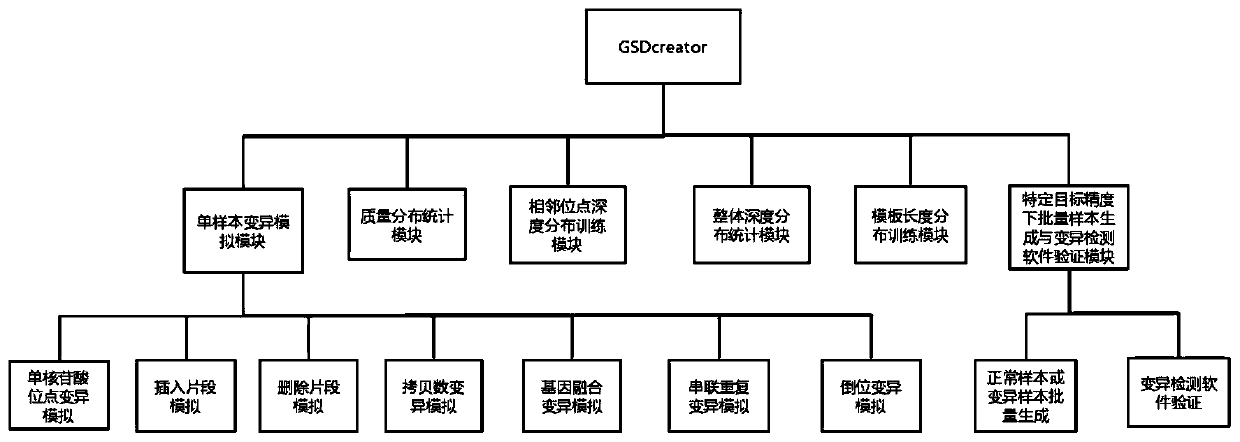
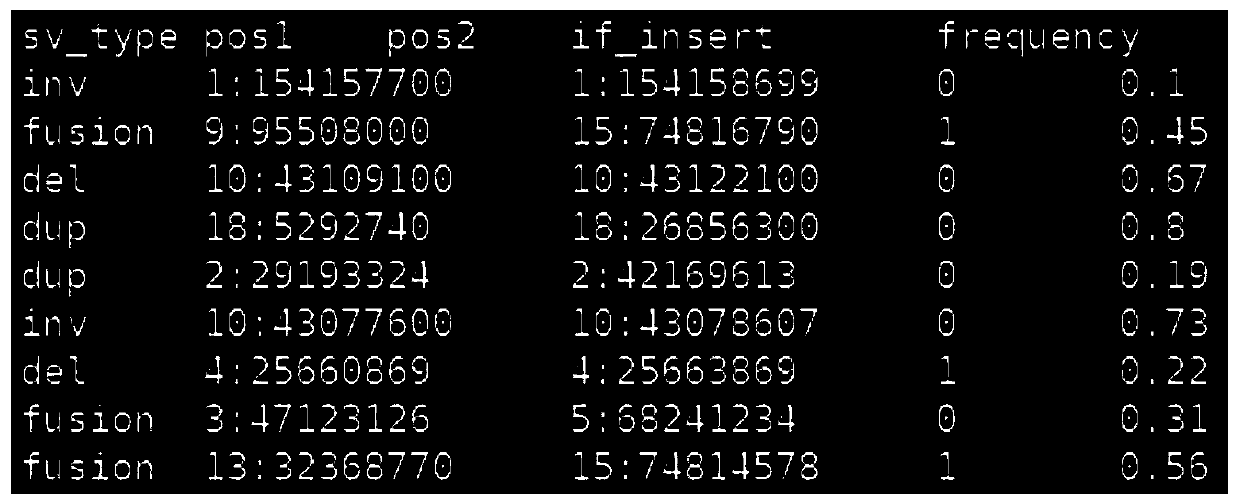
[0076] see figure 1, the present invention is a gene sequencing data simulation method for simulating the background information of a crowd. First, the reference genome file and the target capture area file are loaded; the target capture area file records the start of each target area that the user pays attention to on the reference genome Coordinates and end coordinates; after the file is loaded, the system starts to enter each variation simulation module, according to the sequencing depth set by the user, seven types of variation (single nucleotide site variation, insertion mutation, deletion mutation, copy number variation, Inversion variation, gene fusion variation, tandem duplication variation), variation frequency, and the coordinates of the variation on the reference genome complete the corresponding simulation; at this time, the print station of the program displays a percentage, indicating the running progress of the program; the sequencing file simulation is completed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com