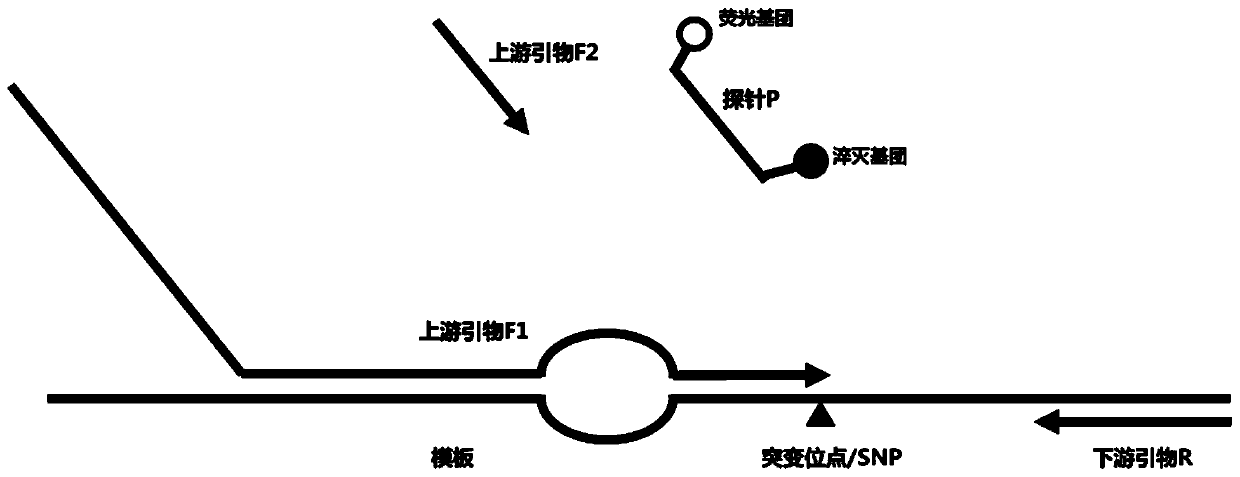
Nucleic acid sequence detection composition and detection method
A nucleic acid sequence and target nucleic acid sequence technology, applied in the field of molecular biology, can solve the problems of increasing detection cost and operational complexity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
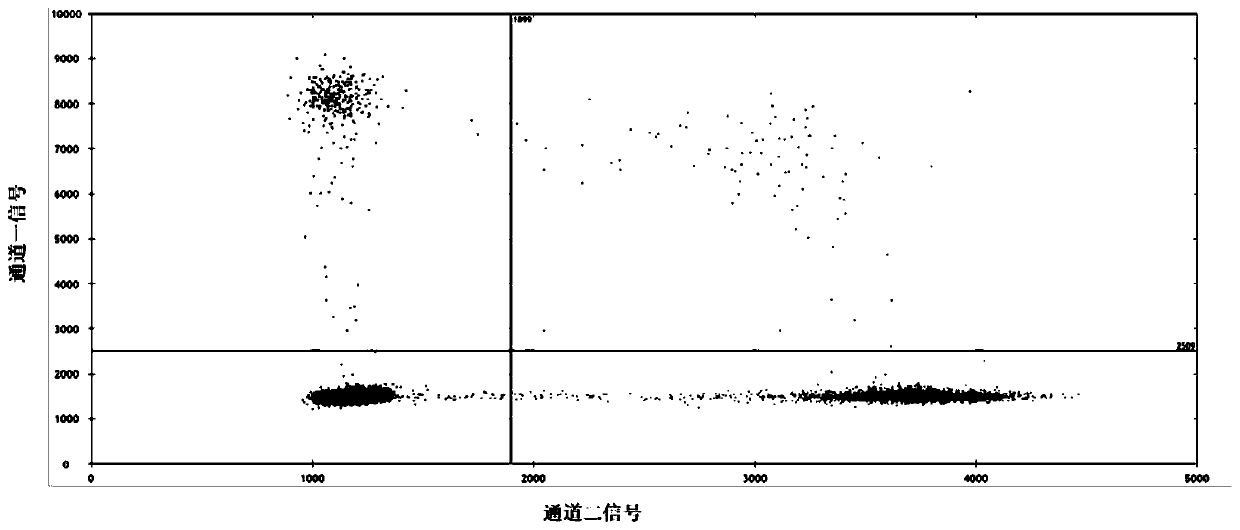
[0078] In this embodiment, a common nucleic acid variation is taken as an example to test the primer probe composition and detection system of the present invention. Specifically, the L858R mutation of the human EFGR gene was used as an example to simulate clinical samples to evaluate the performance of the detection system of the present invention.
[0079]EFGR is a common driver gene of non-small cell lung cancer, and its mutations mainly occur in exons 18 / 19 / 20 / 21, and more than 45% of EGFR driver mutations are encoded by the 858th amino acid of exon 21 from leucine Acid (L) is changed to arginine (R), and the above mutations cause continuous activation of EGFR downstream pathways, leading to tumorigenesis. At the same time, in the process of targeted therapy, tumors caused by the EGFR gene L858R mutation were found to be sensitive to the first-generation EGFR tyrosine kinase inhibitor (EGFR-TKI) drugs, so quantitative detection of EGFR gene L858R mutation and Continuous f...
Embodiment 2
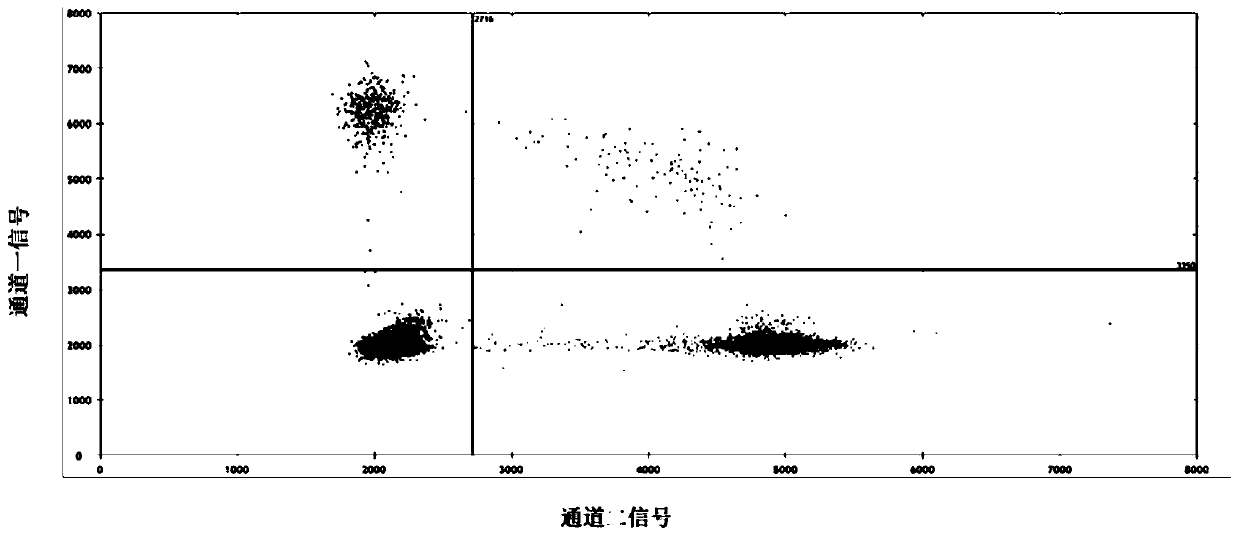
[0113] In order to further verify the effects of the primers and methods of the present invention, another gene variation (human BRAF gene V600E mutation) was detected in this embodiment.
[0114] 1. Sample preparation:
[0115] Fragmented Colo 205 cell line DNA samples quantified by digital PCR, containing the BRAF gene V600E mutation with a mutation abundance of 65.8%, were used to simulate clinical circulating tumor DNA.
[0116] Specifically, with QIAGEN The DNA Mini Kit was used to extract the nucleic acid of the Colo 205 cell line containing the BRAF gene V600E mutation according to the operating instructions of the kit to obtain the genomic DNA of the BRAF gene V600E mutant cell line. Using KAPA Frag Kit, the extracted BRAF gene V600E mutant cell line DNA was digested and fragmented to obtain fragmented mutant DNA with a fragment length of about 120-130bp, which simulated the fragment size of clinical circulating tumor DNA.
[0117] The free DNA extracted from the pl...
Embodiment 3
[0143] In this embodiment, the DNA sample of NCI-H1650 cell line was used to test the detection effect of the primer probe composition and the detection method of the present invention on the deletion mutation for exon 19 mutation of human EGFR gene.
[0144] 1. Sample preparation:
[0145] The 293T cell line DNA purified by ultrasound was prepared, and it was confirmed by next-generation sequencing that it did not contain the EGFR gene 19del mutation, and it was used as a negative sample.
[0146] At the same time, the NCI-H1650 cell line DNA sample quantified by digital PCR was prepared. After fragmentation, the wild-type DNA sample was used to dilute the EGFR gene 19del mutation sample to a mutation abundance of 10% as a positive sample.
[0147] The blank control was Tris-EDTA buffer without DNA.
[0148] 2. Preparation of reaction system
[0149] 2.1 Primer probe composition
[0150] The primers and probes of the present invention are synthesized by Sangon Bioengineeri...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com