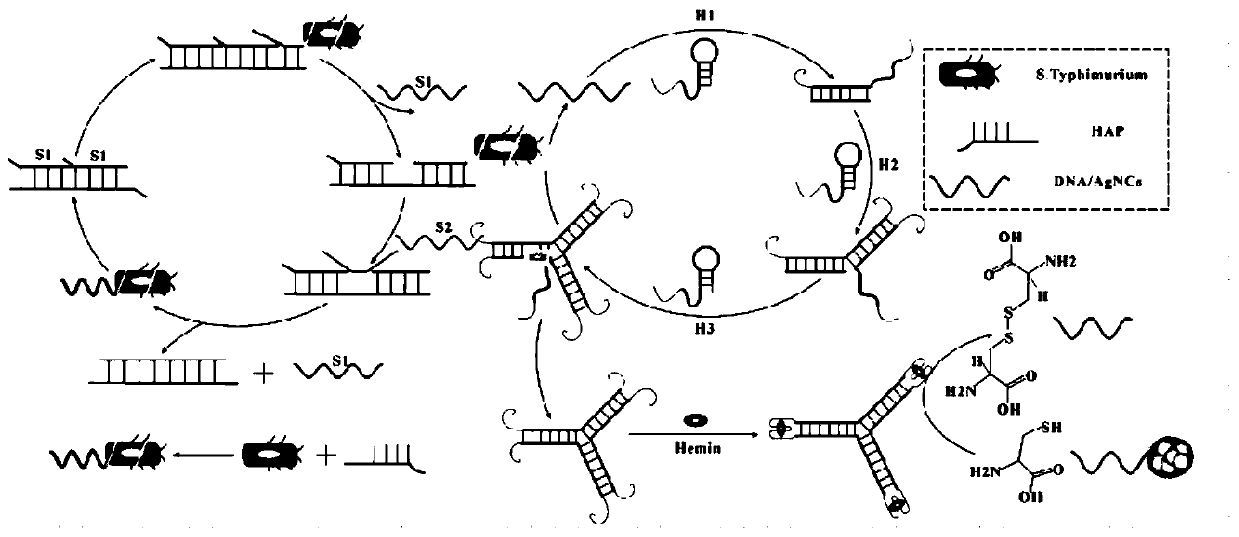
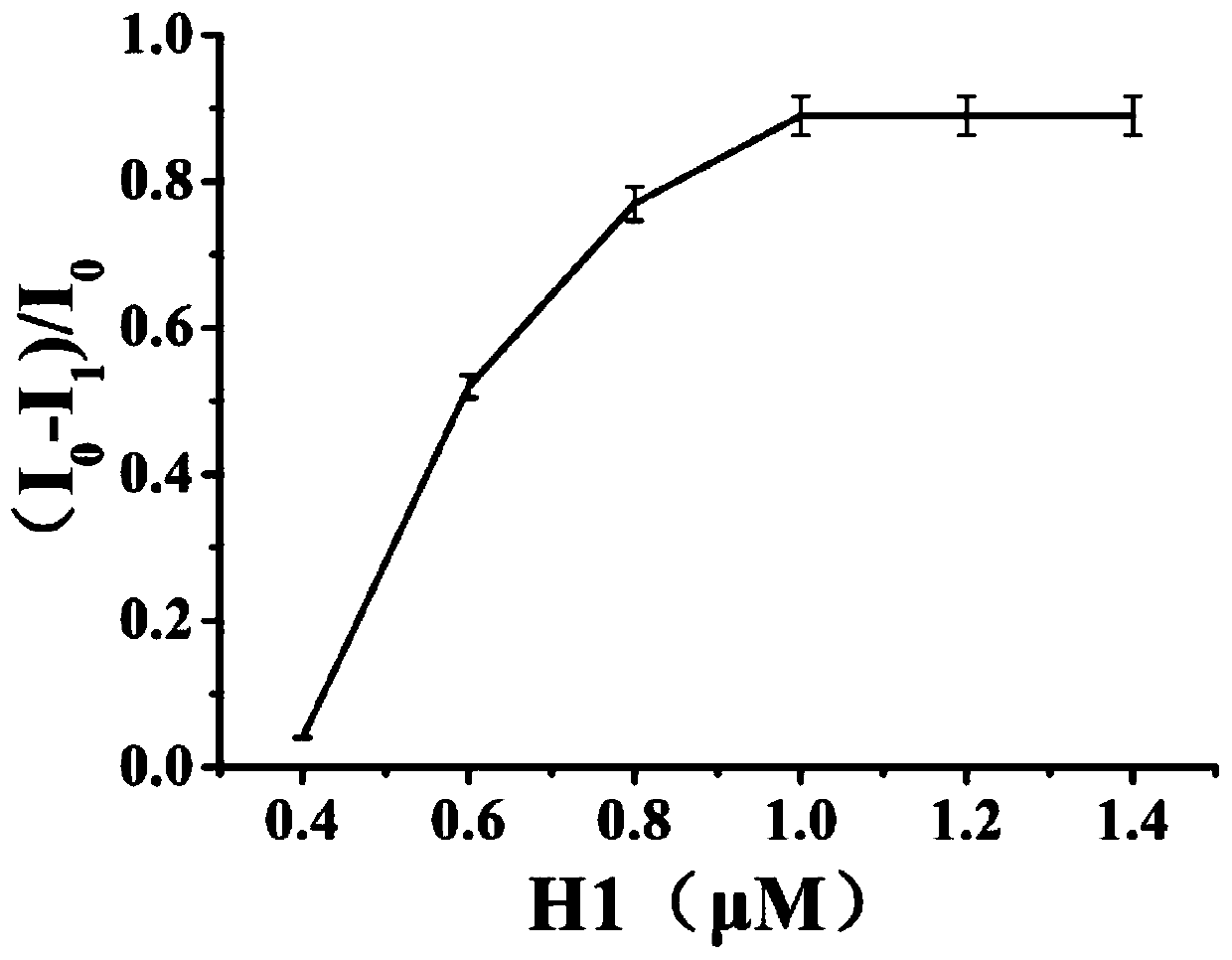
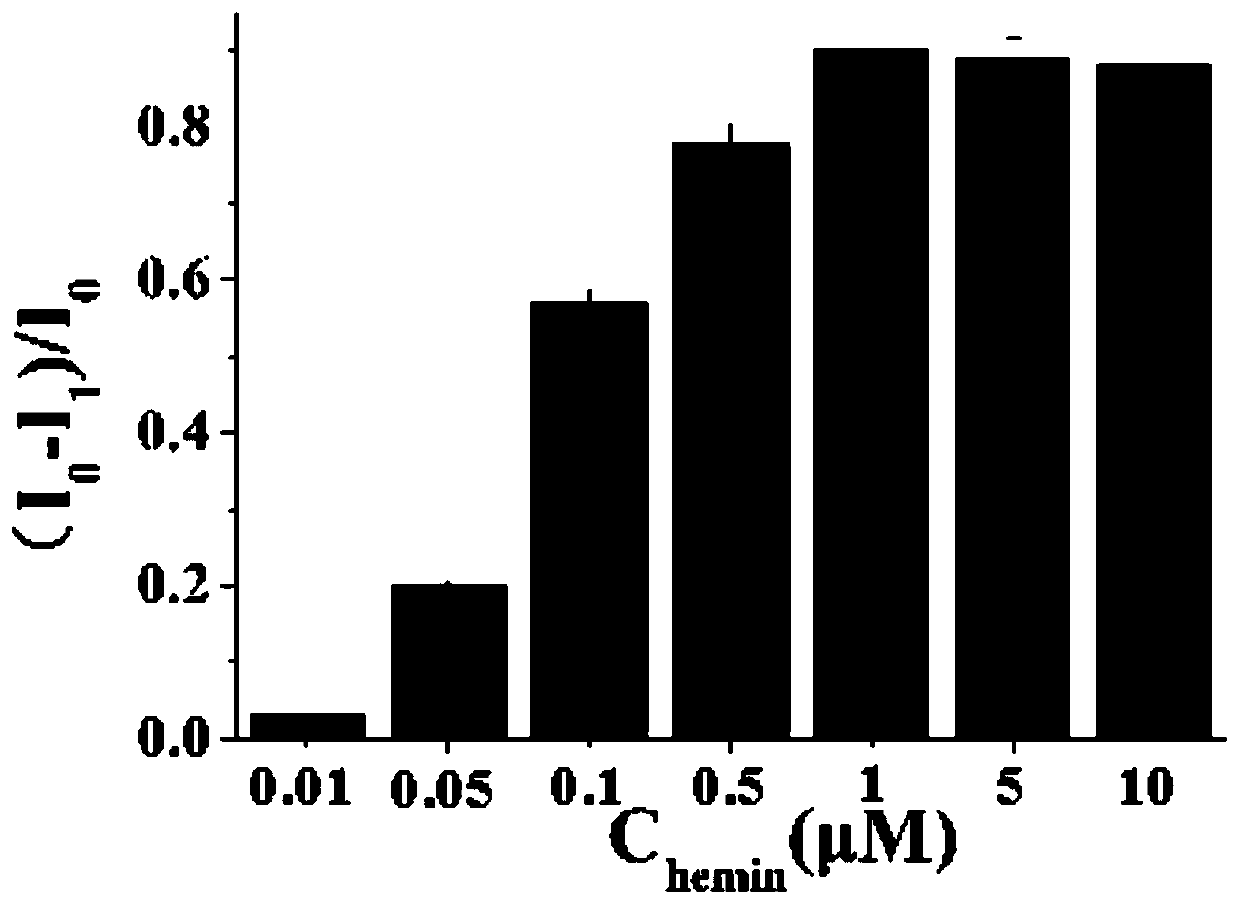
Biosensor for detecting salmonella based on aptamer and preparation method thereof and application
A biosensor, Salmonella technology, applied in the field of biosensors, can solve the problems of long detection period, high cost, low specificity and sensitivity, etc., and achieve the effects of short detection period, simple operation and improved sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] The preparation method of described fluorescent biosensor comprises the following steps:
[0063] The synthetic operation steps of composite probe S are as follows:
[0064] Add 18 μL of sterilized water, 3 μL of 10×PB, 3 μL of 100 μM S0 probe and 6 μL of 100 μM S1 probe into the pre-prepared sterilized EP tube, shake for 30 seconds, and incubate at 95°C Incubate for 5 minutes, slowly cool down to room temperature and hybridize into probes, and store at -20°C for later use.
[0065] The steps for the synthesis of DNA silver nanoclusters (AgNCs-DNA) are as follows:
[0066] Add 15 μL of 100 μM Nucleic Acid Strand C and 73 μL of 20 mM PB (pH 7.0) buffer into the EP tube wrapped in foil paper, and then add 6 μL of 1.5 mM AgNO3 solution (to ensure the ratio of Ag+ to H3 6:1), shake for 1 min, and place at 4 °C for 30 min; after 30 min, continue to add 6 μL of 1.5 mM NaBH4 to the EP tube, shake for 1 min, and place at 4 °C in the dark for more than 6 h. ;
[0067] The ma...
Embodiment 2
[0074] The preparation method of described fluorescent biosensor comprises the following steps:
[0075] The synthetic operation steps of composite probe S are as follows:
[0076] Add 18 μL of sterilized water, 3 μL of 10×PB, 3 μL of 100 μM S0 probe and 6 μL of 100 μM S1 probe into the pre-prepared sterilized EP tube, shake for 30 seconds, and incubate at 95°C Incubate for 5 minutes, slowly cool down to room temperature and hybridize into probes, and store at -20°C for later use.
[0077] The steps for the synthesis of DNA silver nanoclusters (AgNCs-DNA) are as follows:
[0078] Add 15 μL of 100 μM Nucleic Acid Strand C and 73 μL of 20 mM PB (pH 7.0) buffer into the EP tube wrapped in foil paper, and then add 6 μL of 1.5 mM AgNO3 solution (to ensure the ratio of Ag+ to H3 6:1), shake for 1 min, and place at 4 °C for 30 min; after 30 min, continue to add 6 μL of 1.5 mM NaBH4 to the EP tube, shake for 1 min, and place at 4 °C in the dark for more than 6 h. ;
[0079] The ma...
Embodiment 3
[0086] The preparation method of described fluorescent biosensor comprises the following steps:
[0087] The synthetic operation steps of composite probe S are as follows:
[0088] Add 18 μL of sterilized water, 3 μL of 10×PB, 3 μL of 100 μM S0 probe and 6 μL of 100 μM S1 probe into the pre-prepared sterilized EP tube, shake for 30 seconds, and incubate at 95°C Incubate for 5 minutes, slowly cool down to room temperature and hybridize into probes, and store at -20°C for later use.
[0089] The steps for the synthesis of DNA silver nanoclusters (AgNCs-DNA) are as follows:
[0090] Add 15 μL of 100 μM Nucleic Acid Strand C and 73 μL of 20 mM PB (pH 7.0) buffer into the EP tube wrapped in foil paper, and then add 6 μL of 1.5 mM AgNO3 solution (to ensure the ratio of Ag+ to H3 6:1), shake for 1 min, and place at 4 °C for 30 min; after 30 min, continue to add 6 μL of 1.5 mM NaBH4 to the EP tube, shake for 1 min, and place at 4 °C in the dark for more than 6 h. ;
[0091] The ma...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com