Human SGIP1, SCAND3 and MYO1G gene methylation detection kit
A detection kit and methylation technology, applied in the biological field, can solve the problems of reducing the specificity of sequence hybridization, easy degradation of samples, and reducing detection sensitivity, so as to reduce the occurrence of false positive results, the detection results are accurate and reliable, and the detection efficiency is improved. The effect of sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Preparation of human SGIP1, SCAND3 and MYO1G gene methylation detection kit (fluorescent PCR method) of the present invention
[0049] Human SGIP1, SCAND3 and MYO1G gene methylation detection kit (fluorescent PCR method), including methylation-sensitive restriction endonuclease, digestion buffer, fluorescent PCR reaction solution, negative reference and positive reference. The preparation of the kit includes the following steps:
[0050] (1) Preparation of methylation-sensitive restriction enzymes: select three methylation-sensitive restriction enzymes according to the target gene, namely AciⅠ, BstUⅠ and HpaⅡ, and mix them at a volume ratio of 1:1:1 Prepare an enzyme mixture with a final concentration of 3.3±0.03U / μl for each enzyme, and save it for future use. The preparation scheme of the enzyme mixed solution is as follows:
[0051] Reagent name per 6 reactions (μl) AciⅠ(10U / μl) 1 BstUⅠ(10U / μl) 1 HpaⅡ(10U / μl) 1 total capacity 3 ...
Embodiment 2
[0072] Methylation-sensitive restriction endonuclease verification experiment of the present invention
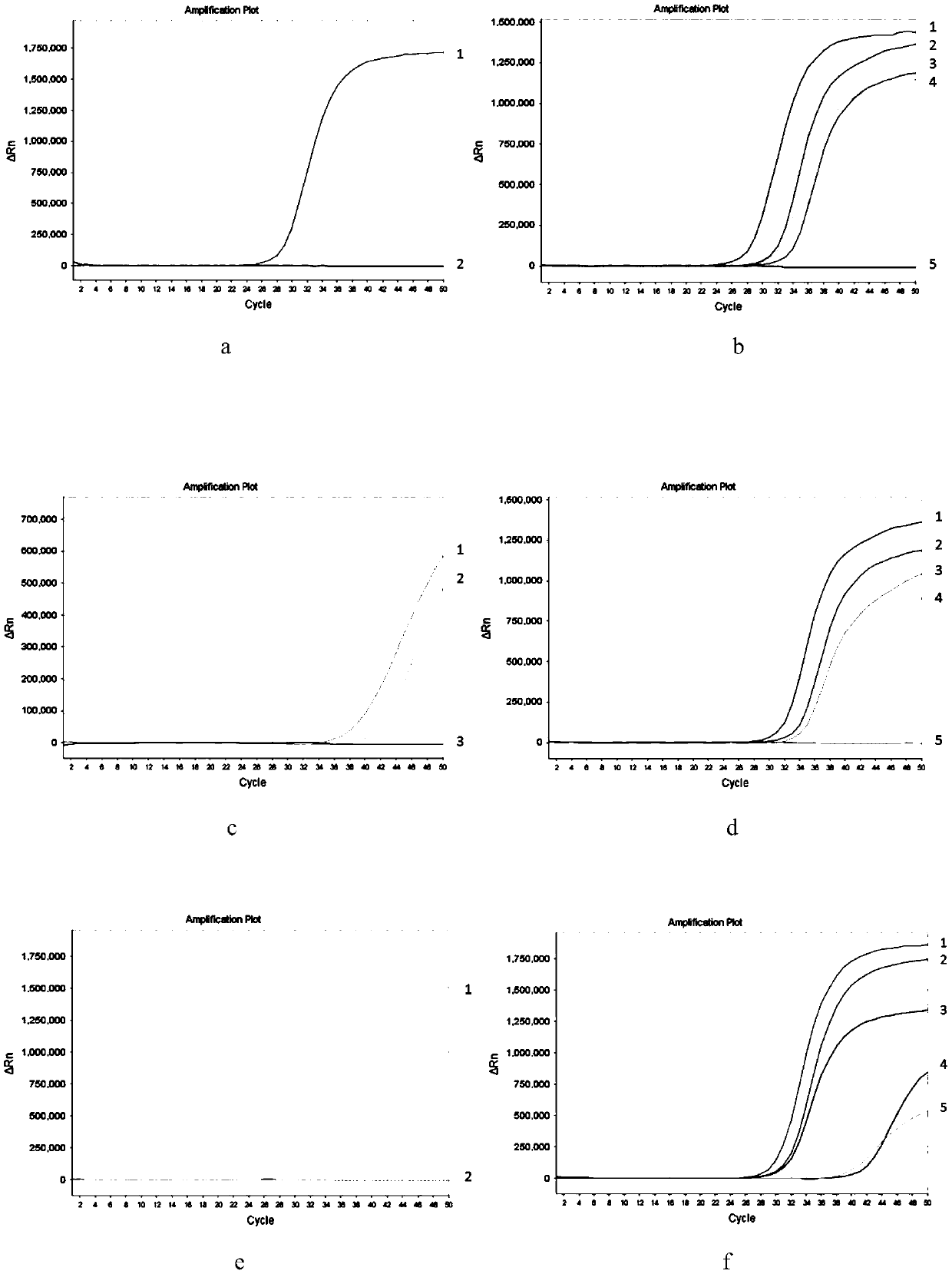
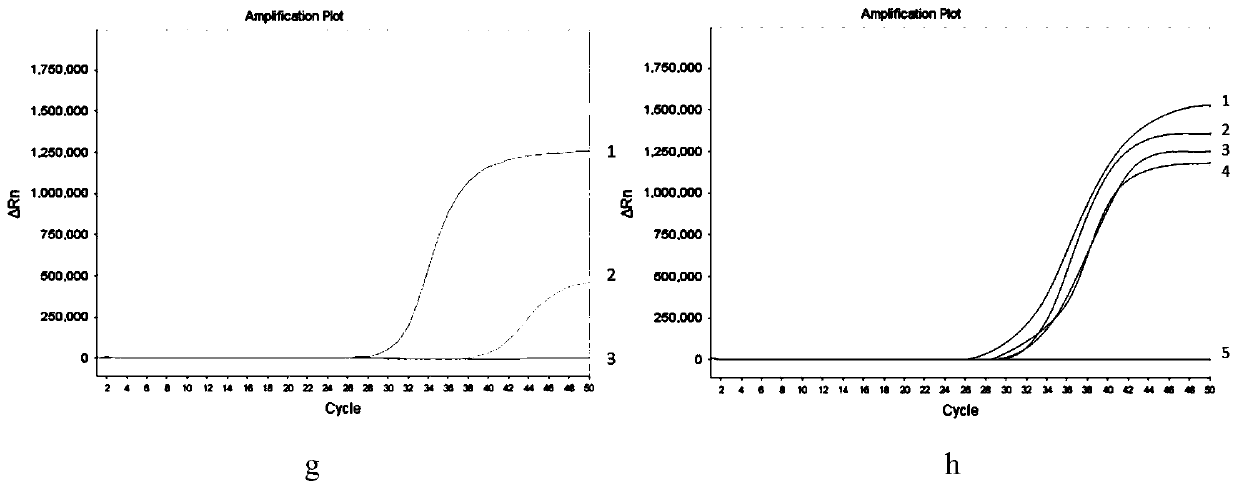
[0073] (1) Purpose of the experiment
[0074] In this example, the enzymatic cutting effect of the methylation-sensitive restriction enzyme of the present invention is verified by comparing with different methylation-sensitive restriction enzymes.
[0075] (2) Experimental method
[0076]In this example, non-methylated human genomic DNA, methylated human genomic DNA of SGIP1, SCAND3 and MYO1G determined by bisulfite sequencing method were selected as samples to be tested, and the methylated human genomic DNA in the kit described in Example 1 was used respectively The enzyme-sensitive restriction endonucleases (abbreviated as the present invention in the following table) and AciI, BstUI, and HpaII carried out enzyme digestion reactions, each of which was repeated 3 times, and the total amount of DNA added in the enzyme digestion reaction system remained consistent. The enz...
Embodiment 3
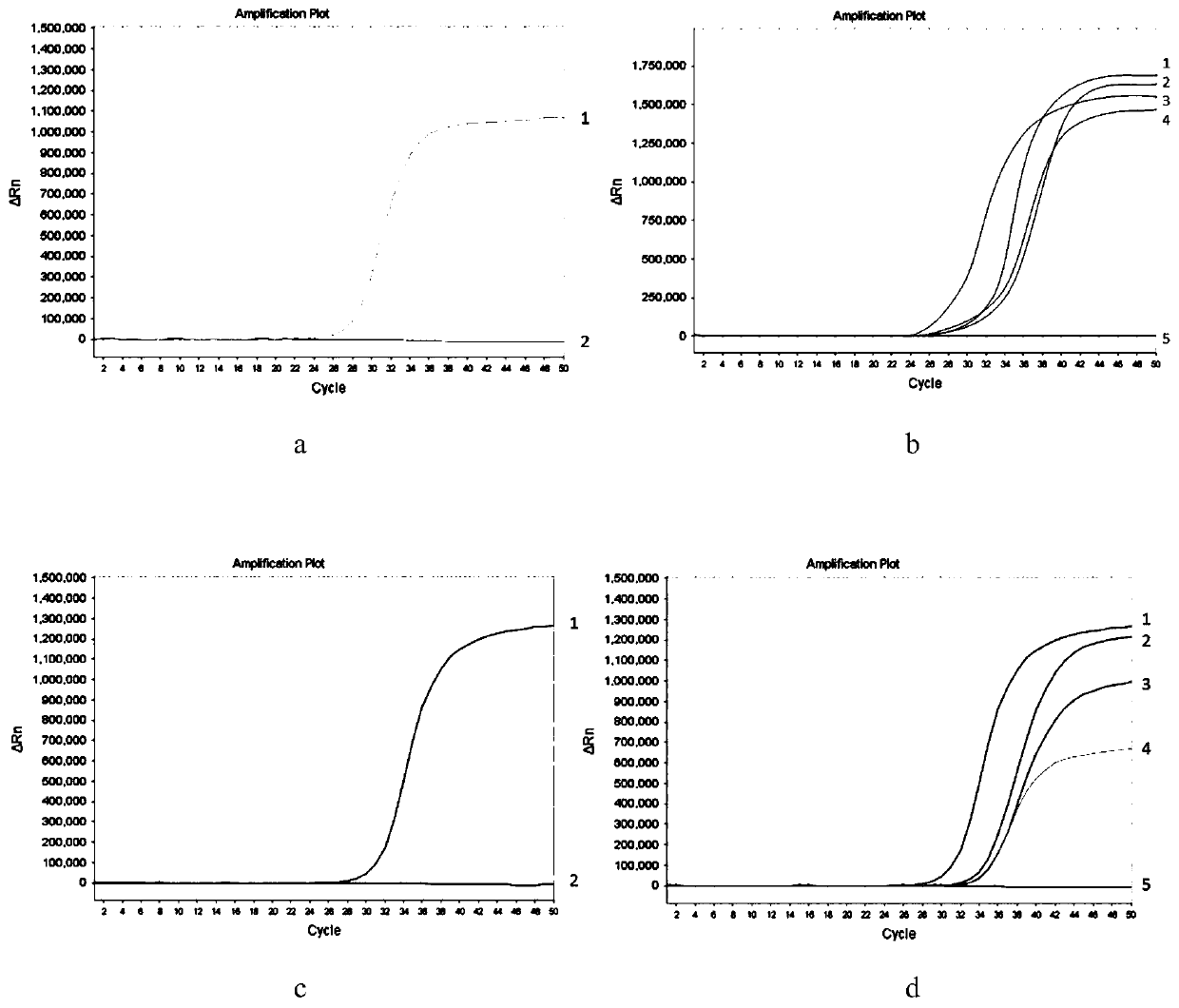
[0083] Sample pretreatment method of the present invention compares with bisulfite conversion method
[0084] (1) Purpose of the experiment
[0085] In this example, the method for detecting methylated DNA based on methylation-sensitive restriction endonuclease digestion and the method for detecting methylated DNA based on bisulfite conversion of the present invention were compared.
[0086] (2) Experimental method
[0087] In this example, non-methylated human genomic DNA, methylated human genomic DNA of SGIP1, SCAND3 and MYO1G determined by bisulfite sequencing method were selected as samples to be tested, and the methylation-sensitive restriction endogenous DNA of the present invention was used respectively Dicer and bisulfite were used to pre-treat the DNA samples to be tested, with 3 replicates each. The methylation-sensitive restriction endonuclease digestion reaction of the present invention is operated according to the detection steps in Example 1; bisulfite conversi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com