Method for efficiently expressing hypersensitive protein by using T4 phage display technology
A phage display and hypersensitive protein technology, applied in the field of high-efficiency expression of hypersensitive proteins, can solve the problems of lengthy process, reduced yield of microbial proteins and polypeptides, and many steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1. Sequence acquisition of hypersensitive protein (harpinEa)
[0029] The sequence of gene hrpN encoding harpinEa protein derived from strain Erwinia amylovora has been published (GeneBank number: M92994.3). The hypersensitive protein (harpinEa) gene can be obtained using well-known techniques in the industry such as gene synthesis, PCR amplification, extraction of chromosomal DNA and establishment of a library. The present invention adopts the gene synthesis method to send the hypersensitive protein (hrapinEa) amino acid sequence to be synthesized (Suzhou Jinweizhi Biotechnology Co., Ltd., China).
Embodiment 2
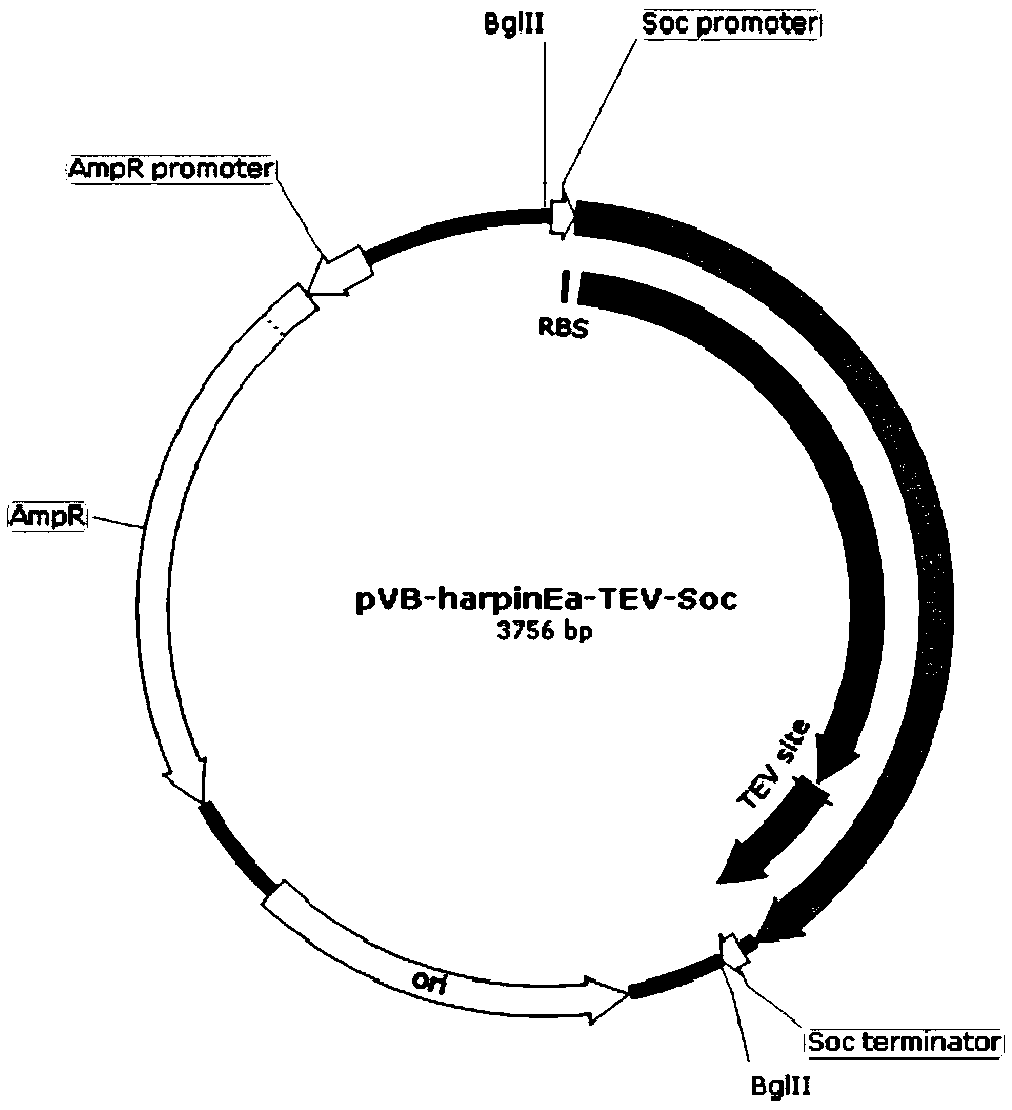
[0030] Embodiment 2, construct pVB-harpinEa-TEV-Soc plasmid
[0031] The backbone of the plasmid was from the pUC18 plasmid. Using the pUC18 plasmid as a template, the ampicillin resistance gene and the plasmid replication region were amplified by PCR using KOD high-fidelity DNA polymerase. The forward primer of PCR is 18 bases in the upstream sequence of pUC18 ampicillin resistance gene, and the reverse primer is the reverse complementary sequence of 18 bases in the downstream sequence of pUC18 plasmid replication region. A BglII restriction enzyme site was added to the 5' ends of the forward and reverse primers. At the same time, we used T4 phage DNA as a template and obtained the Soc protein gene fragment by PCR amplification using specific primers, wherein the 5' end of the forward primer introduced the gene sequence of TEV protease. The synthetic harpinEa protein gene fragment and the TEV-Soc protein gene fragment were fused into a DNA molecule by bridging PCR, wherein ...
Embodiment 3
[0033] Example 3. In vivo expression and phage display of fusion recombinant protein
[0034] The T4 phage synchronous display expression plasmid pVB-harpinEa-TEV-Soc was transformed into competent cells of E. coli P301 strain by chemical transformation method, and under the pressure of ampicillin antibiotics, it was cultured in a shaker at 37°C at a speed of 200rpm until the bacteria Density up to 3×10 8 pieces / ml. Soc with Soc protein deletion - T4 phage (gifted by the laboratory of Professor Rao, Department of Biology, Catholic University of America) infected the cultured Escherichia coli cells at MOI (multiplicity of infection) = 1, and continued to culture in a shaker at 37° C. at 200 rpm for 30 min. During this process, the harpinEa-TEV-Soc recombinant protein was expressed under the regulation of the T4 phage particle assembly pathway and displayed on the capsid.
[0035] After the display is completed, the culture solution is immediately divided into pre-cooled centri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com