Rice drought tolerance-related transcription factor OsAE10 and coding gene and application thereof
A rice transcription factor and gene technology, applied in the application, genetic engineering, plant gene improvement and other directions, can solve the problem that the function of the member has not been identified.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
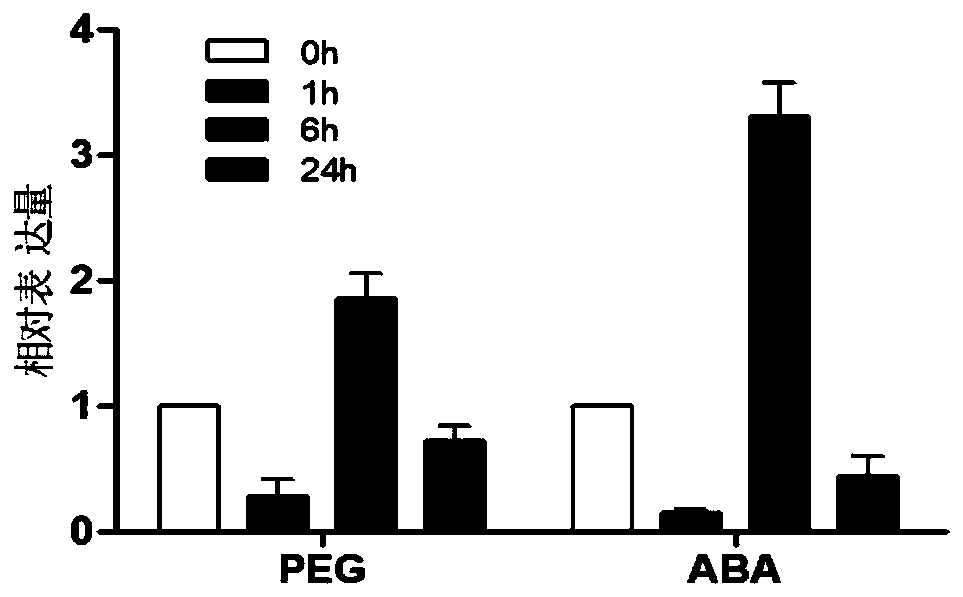
[0036] Example 1 The expression of OsAE10 gene is induced by drought and ABA
[0037] 1. Analysis of the expression pattern of OsAE10 gene under different treatment conditions
[0038] The wild-type rice was treated with 20% PEG and 5 μM ABA respectively, and the leaves at 0h, 1h, 6h and 24h were taken as materials, and the total RNA of different materials was extracted with Trizol, and quantitative PCR analysis was performed after reverse transcription to detect Transcript levels of OsAE10. The primer sequences used to detect the OsAE10 gene are as follows: forward primer: 5′-TCAACTTCCCTTCCGCTACCAC-3′; reverse primer: 5′-ACCTCCTCTCGTCCTCCTCCT-3′. The primers used to detect the expression level of the internal reference gene (actin) were: forward primer: 5'-TGGCATCTCTCAGCACATTCC-3'; reverse primer: 5'-TGCACAATGGATGGGTCAGA-3'. The expression patterns of OsAE10 gene under different treatment conditions are shown in figure 1 . The results showed that when the OsAE10 gene was...
Embodiment 2
[0039] Example 2 Cloning of OsAE10 gene and analysis of transcription factor characteristics of its encoded protein
[0040] 1. Cloning of OsAE10 gene
[0041] Design the PCR primer sequence, the forward primer is: 5'-GCAAAAGAAAAGCAGCAAC-3', the reverse primer is: 5'-AAGGAACCAGGACGAACAC-3', using the cDNA of the drought stress 6h used in the quantitative experiment in Example 1 as a template, according to Conventional PCR procedure, RT-PCR amplification, electrophoresis of the amplified product, recovery, addition of A, transformation of Escherichia coli DH-5α after connection with T-carrier, screening of positive clones and sequencing, the clones obtained with correct sequencing were named as pT-OsAE10, completed gene cloning.
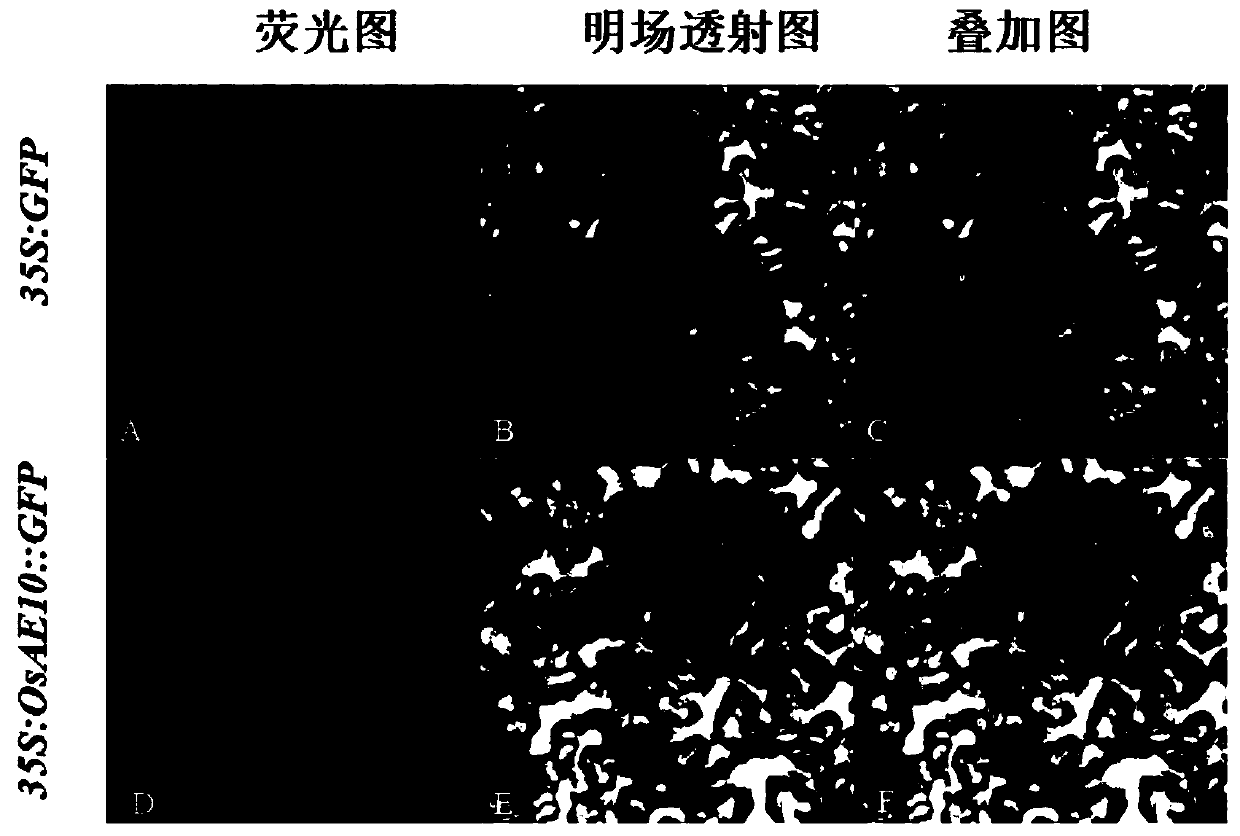
[0042] 2. OsAE10 protein is localized in the nucleus
[0043]The fusion expression vector p1300-OsAE10::GFP of OsAE10 and GFP was constructed, and the p1300-OsAE10::GFP plasmid and the control p1300-GFP were transformed into Agrobacterium, respectiv...
Embodiment 3
[0047] Example 3 Identification of Drought Tolerance Function of OsAE10 Gene
[0048] The pMDC32-OsAE10 overexpression vector was constructed and transformed into Agrobacterium EHA105; after the correct transformants were obtained, they were respectively transformed into Nipponbare rice to obtain transgenic rice plants. The DNA and RNA levels of the obtained different transgenic rice lines were identified ( Figure 4 ) and cultivated for 3-4 generations to obtain transgenic homozygous lines for the following experiments.
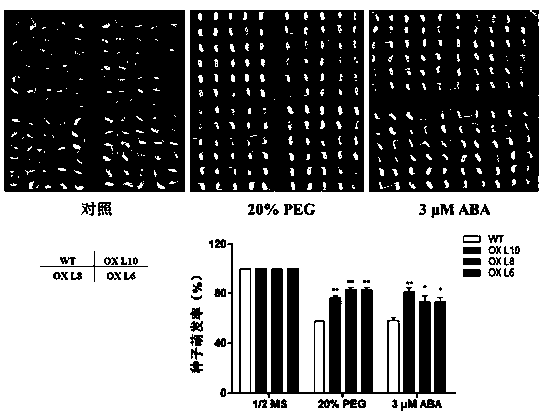
[0049] 1. Detection of germination rate of transgenic rice seeds overexpressing OsAE10 under stress conditions
[0050] The seeds of the wild type and different OsAE10 overexpression strains were sterilized and planted on the medium of 1 / 2MS0, 1 / 2MS+20%PEG and 1 / 2MS+3μM ABA for germination, and the germination rate was counted. It was found that on the 1 / 2MS0 medium, the germination rates of seeds of wild-type rice and OsAE10 overexpression lines were basi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com