DNA polymerase mutant with improved thermal stability, and construction method and application thereof
A thermal stability and mutant technology, applied in the field of DNA polymerase mutants and their construction, can solve the problems of poor thermal stability of DNA polymerase, etc., and achieve the effects of short culture period, good application prospects and simple culture conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
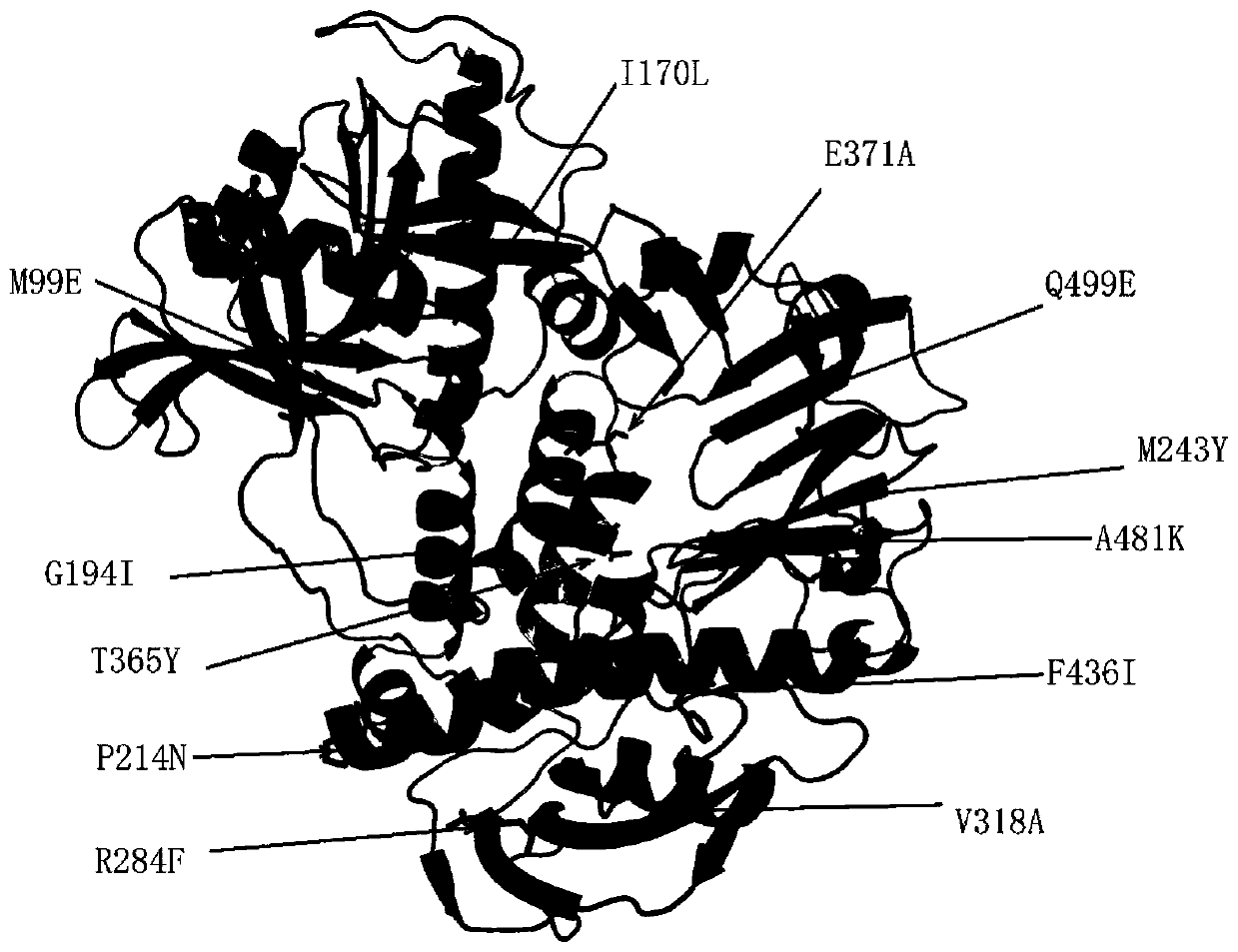
[0039] This embodiment provides a DNA polymerase mutant with improved thermostability, which is a molecularly engineered derivative protein based on the amino acid sequence of the wild-type DNA polymerase (AAA32368.1), wherein the wild Type DNA polymerase (AAA32368.1) is a protein expressed by the wild-type DNA polymerase (AAA32368.1) gene derived from Bacillus phage M2 after codon optimization, and the nucleic acid sequence encoding the protein It is SEQ ID NO.1, and the amino acid sequence is SEQ ID NO.2.
[0040] The DNA polymerase mutants with improved thermostability provided in this example include:
[0041] (a1) a derivative protein having the same function as the protein shown in SEQ ID NO.2 by substituting, deleting or adding one or more amino acids to the amino acid sequence shown in SEQ ID NO.2; or
[0042] (a2) A derivative protein that substitutes one or more amino acid residues at one or more positions of the amino acid sequence shown in SEQ ID NO.2 and exhibits...
Embodiment 2
[0060] The present embodiment provides a gene encoding the DNA polymerase mutant as described in Example 1:
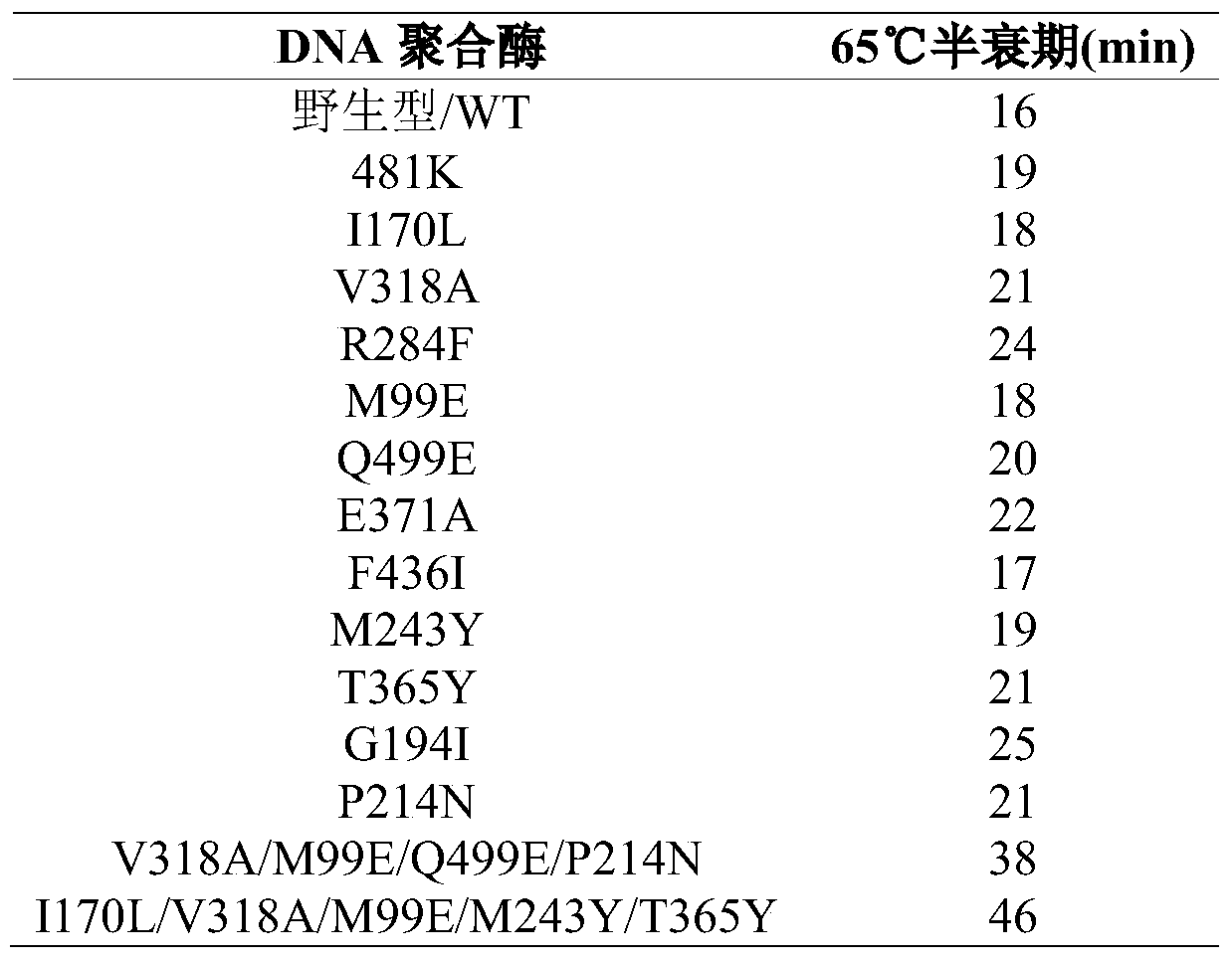
[0061] (1) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is A481K is SEQ ID NO.3;
[0062] (2) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is I170L is SEQ ID NO.4;
[0063] (3) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is V318A is SEQ ID NO.5;
[0064] (4) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is R284F is SEQ ID NO.6;
[0065] (5) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is M99E is SEQ ID NO.7;
[0066] (6) The nucleic acid sequence encoding the DNA polymerase single-point mutant whose mutation site is Q499E is SEQ ID NO.8;
[0067] (7) The nucleic acid sequence encoding the DNA polymerase single-point mutant wh...
Embodiment 3
[0076] This embodiment provides a recombinant plasmid comprising the gene provided in Example 2, which is a plasmid vector that can be used to integrate DNA polymerase single point mutants and combined mutant genes into host cells for stable expression, such as pET28a, pET20b, pET9a, pUC18, pUC19, pET15b, pET22a, etc., can also be used to integrate DNA polymerase genes into host cells for expression; host cells can be selected from Gram-negative bacteria (such as E. coli), Gram-positive bacteria (such as Bacillus subtilis), fungi (such as Aspergillus), yeast (such as Saccharomyces cerevisiae), actinomycetes (such as Streptomyces), etc., can be used to express DNA polymerase single point mutants and combined mutant proteins.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com