Application of single domain antibody for hepatitis a viruses
A hepatitis A virus and single-domain antibody technology, applied in the field of biotechnology or immunology, can solve the problems of high detection cost, large differences in antibody affinity, and poor repeatability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1: Construction of a single-domain antibody library against hepatitis A virus:
[0073] (1) Use 1ml of hepatitis A virus inactivated vaccine to immunize an Inner Mongolia Alxa Bactrian camel, once a week, a total of 7 consecutive immunizations, during which B cells are stimulated to express specific nanobodies; (2) After the immunization Finally, extract 100ml of camel peripheral blood lymphocytes and extract total RNA; (3) synthesize cDNA and use nested PCR to amplify VHH; (4) use restriction endonucleases Pst I and Not I to digest 20 μg pMECS phage display vector and 10 μg VHH and ligated the two fragments; (5) Transformed the ligated product into electroporation-competent cells TG1, constructed a phage display library for the capsid protein of hepatitis A virus and determined the storage capacity, the size of the library storage capacity was about 2×10 9 ; At the same time, the correct insertion rate of the target fragment in the built library was detected by...
Embodiment 2
[0074] Embodiment 2: the expression of hepatitis A virus capsid protein:
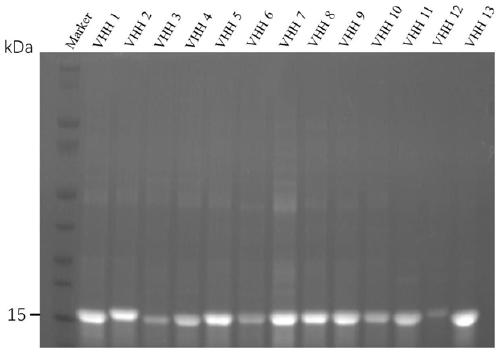
[0075] (1) The sequence information of the hepatitis A virus capsid protein was found from the protein database of NCBI (AccesionNumber: P08617.1); (2) After Uniprot analysis, VP2-VP3-VP1 was selected as the target expression fragment; (3) Add kozak enhancer sequence (5'-GGATCGAACCCTT-3') and IgG kappa signal peptide (METDTLLLWVLLLWVPGSTGD) at the 5' end of VP2-VP3-VP1, and add 6*His tag fusion expression at the 3' end; (4) Examples of use The scheme described in 8 carries out eukaryotic expression; (5) purify with reference to the scheme described in Example 9 using nickel column affinity chromatography; (6) carry out SDS-PAGE analysis with reference to the method of Example 9, and determine that the protein purity is at 95 % or more, and the concentration is more than 0.5mg / mL, keep it at low temperature for later use.
Embodiment 3
[0076] Example 3: Screening for single domain antibodies against hepatitis A virus:
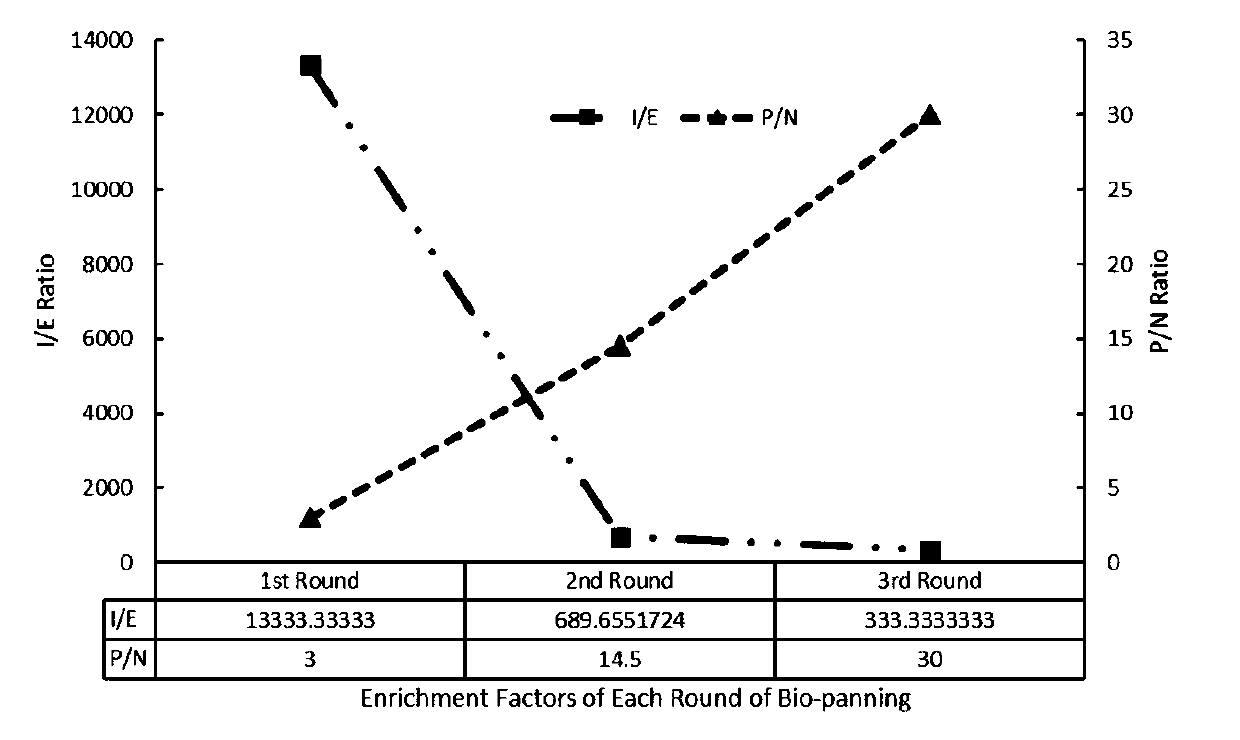
[0077] (1) Take 200 μL of recombinant TG1 cells to culture in 2×TY medium, add 40 μL of helper phage VCSM13 to infect TG1 cells during the period, and culture overnight to amplify phages, use PEG / NaCl to precipitate phages the next day, and centrifuge to collect amplified phages ; (2) NaHCO diluted in 100mM pH 8.3 3 500ug of neutravidin protein in the medium was coupled to the microtiter plate, placed overnight at 4°C, and a negative control well was set up at the same time; (3) the next day, 100 μL of biotin-labeled hepatitis A virus capsid protein (VP2- VP3-VP1-Biotin), incubate at room temperature for 2 hours, add 100 μL PBS to the negative control well; (4) After 2 hours, add 100 μL of 3% skim milk, and block at room temperature for 2 hours; (5) After the end of blocking, add 100 μl amplification Post-phage library (approximately 2×10 11 phage particles) at room temperature for 1 h; (6)...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com