Novel specific molecular target for vibrio parahaemolyticus and rapid detection method thereof
A technology of Vibrio hemolyticus and a primer set, applied in the field of microbial testing, can solve the problems of human health hazards, difficult to meet the actual detection, unable to achieve accurate screening of Vibrio parahaemolyticus, etc., to reduce the detection cost, The results are easy to determine and the specificity is good.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1 Discovery of new molecular targets specific to Vibrio parahaemolyticus
[0046] According to the GenBank database and the whole genome DNA sequence of Vibrio parahaemolyticus self-tested by our team, bioinformatics analysis was performed; 23 non-essential genes unique to Vibrio parahaemolyticus strains were screened, and 20 parahaemolytics were obtained by further analysis and screening Specific gene fragments of Vibrio sex, the nucleotide sequences of the gene fragments are shown in SEQ ID NO. 1-20.
Embodiment 2
[0047] Example 2 Rapid detection method of Vibrio parahaemolyticus
[0048] 1) Primer design
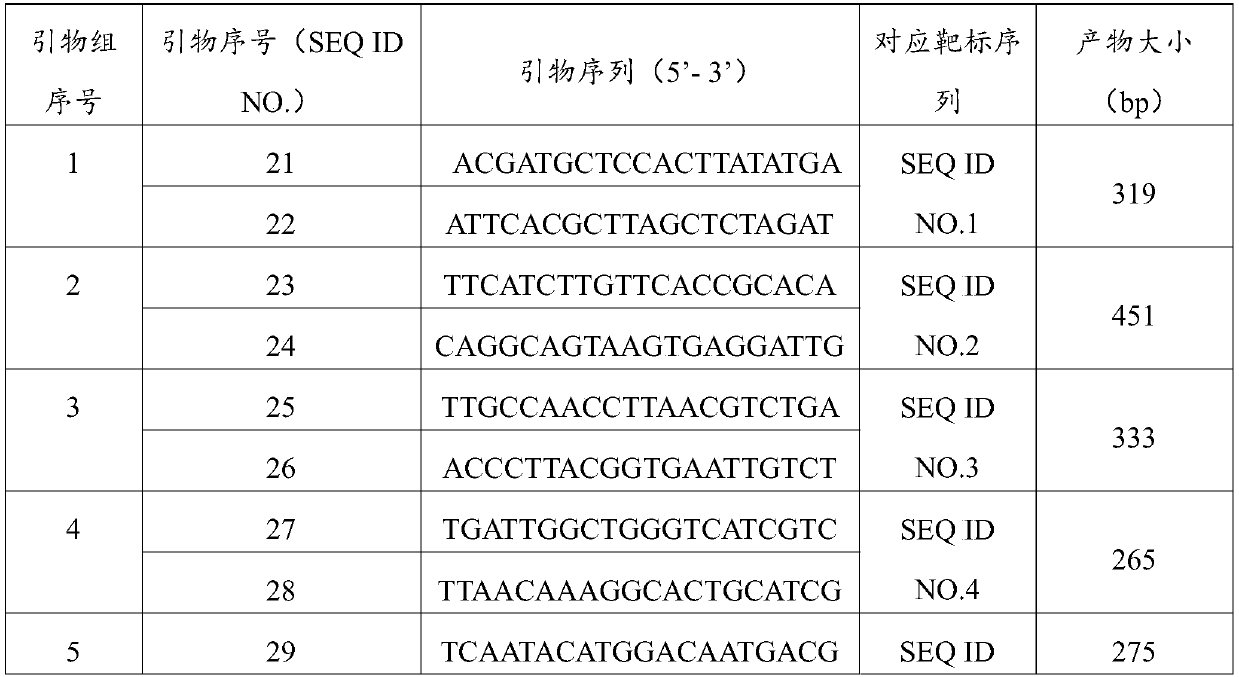
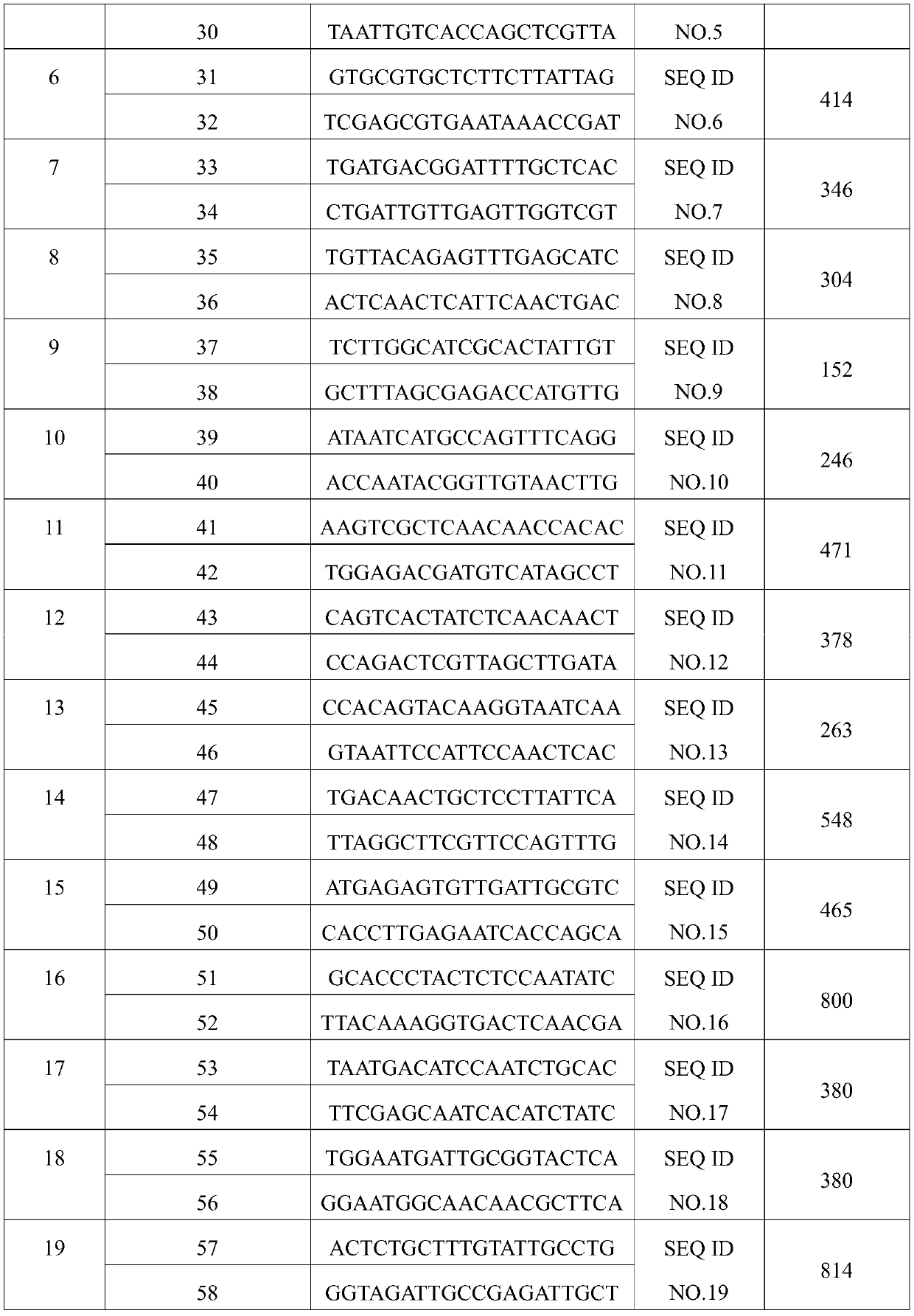
[0049] A specific PCR amplification primer set (including forward primer and reverse primer) was designed according to the sequence SEQ ID NO. 1-20 in Example 1. The sequence of the primer set is shown in Table 1 below.
[0050] Table 1 Specific PCR detection primer set
[0051]
[0052]
[0053]
[0054] 2) The method for identifying Vibrio parahaemolyticus, the steps are as follows:
[0055] S1DNA template preparation: the test strains are respectively enriched and cultured in LB liquid medium, and the bacterial genomic DNA is extracted separately using a commercial bacterial genomic DNA extraction kit as the template to be tested;
[0056] S2PCR amplification: use one of the primer sets 1-20 to perform PCR amplification on the DNA of the test sample
[0057] ①PCR detection system:
[0058]
[0059] ②PCR amplification program:
[0060]
[0061] S3: Take the PCR amplified product and perform gel ...
Embodiment 3
[0063] Example 3 Results of specificity evaluation of PCR detection method
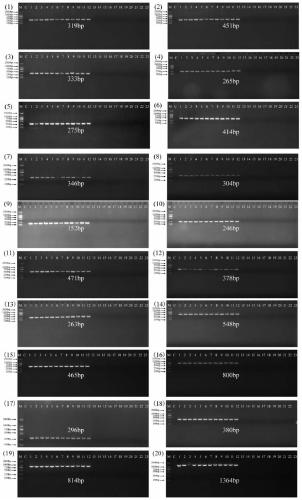
[0064] Twelve strains of Vibrio parahaemolyticus, 2 strains of non-target Vibrio (Vibrio fluvialis and Vibrio vulnificus), 9 strains of non-parahaemolytic Vibrio such as Staphylococcus aureus and Salmonella enteritidis were detected by PCR according to the method of Example 2. , Where the S1 DNA template is prepared by extracting the genomic DNA of each bacteria. Set a blank control. The template of the blank control is an aqueous solution without genome.
[0065] The strains and test results of the bacteria used are shown in Table 2 below. In the table, the "+" in the test result column indicates positive and "-" indicates negative. The results of PCR product electrophoresis are as follows figure 1 As shown; among them, (1) to (20) respectively correspond to the primer sets with serial numbers 1 to 20 in Example 1, M is 2000Maker, C is a blank control, and 1 to 23 respectively correspond to the strains i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com