Gene editing system based on CRISPR-Cas9 technology and application of gene editing system
A gene editing and gene technology, applied in the field of gene editing system, can solve the problems of large base size of plasmid, off-target gene editing, difficult Pf multiple rounds of gene editing, etc., and achieve the effect of reducing use and base size
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
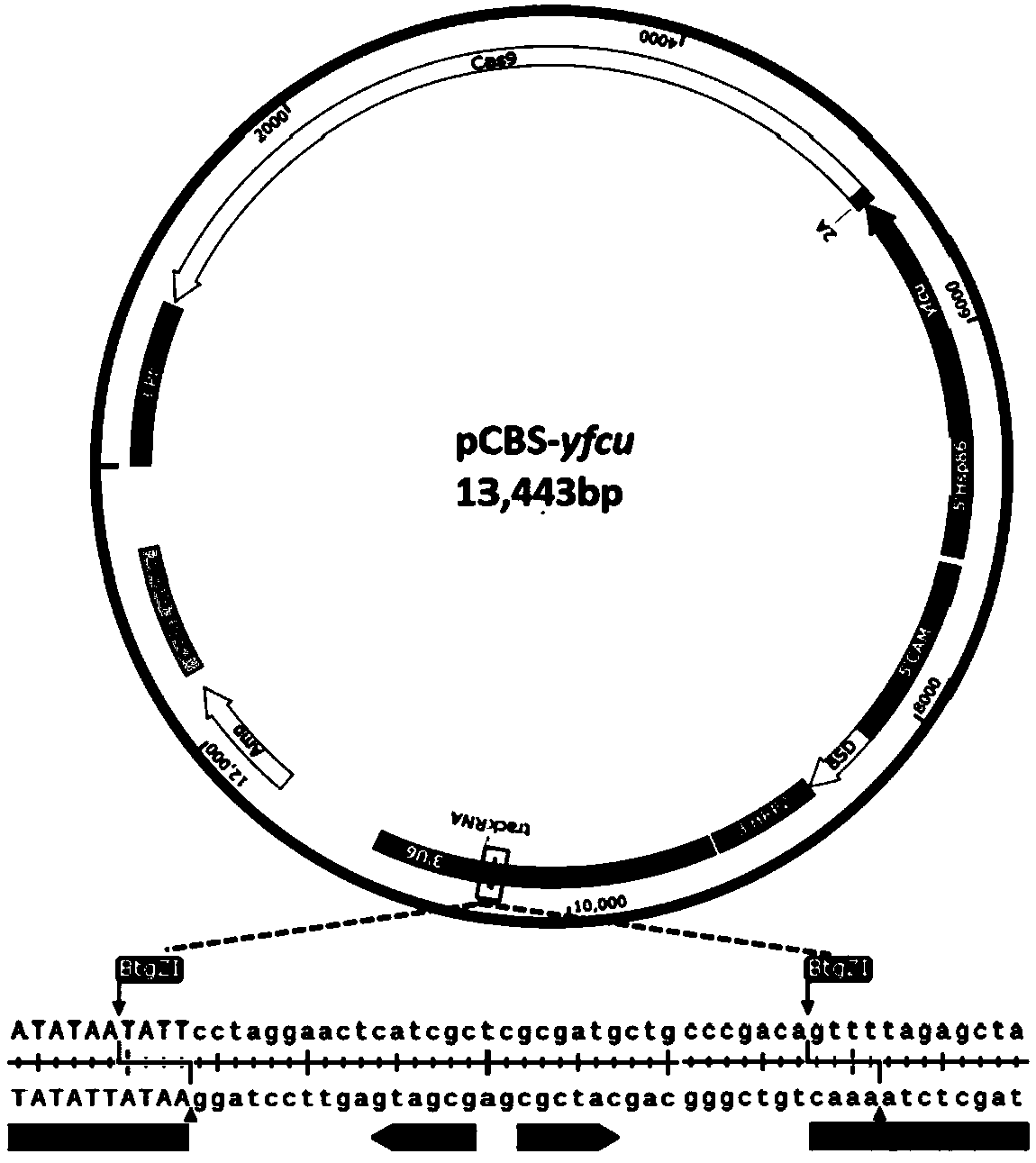
[0086] Example 1 Design and Construction of Universal Cas9 / sgRNA Expression Vector pCBS-yfcu Plasmid
[0087] First, the Cas9 / sgRNA expression vector pCBS plasmid was improved, and the negative selection gene yfcu derived from Saccharomyces cerevisiae was introduced into the vector;
[0088] Since the size of the pCBS plasmid has exceeded 12kb, and a complete negative selection gene expression cassette is inserted, the size will be close to 15kb; in order to reduce the difficulty of vector construction, the yeast-derived negative selection gene yfcu and cas9 gene were incorporated into the same expression cassette, The two are connected by a 2A peptide sequence (SEQ ID NO.2: GGTTCGGGAGAGGGCAGAGGATCCCTGCTAACATGCGGTGATGTCGAGGAGAATC CTGGCCCAGAATCGCTCGAG), and the yfcu-2A fusion gene is directly inserted into the XhoI site before the ORF of the Cas9 gene. The specific construction process is as follows:
[0089] The first step: pCC4 plasmid (Maier, AG., et al. (2006). "Negative se...
Embodiment 2
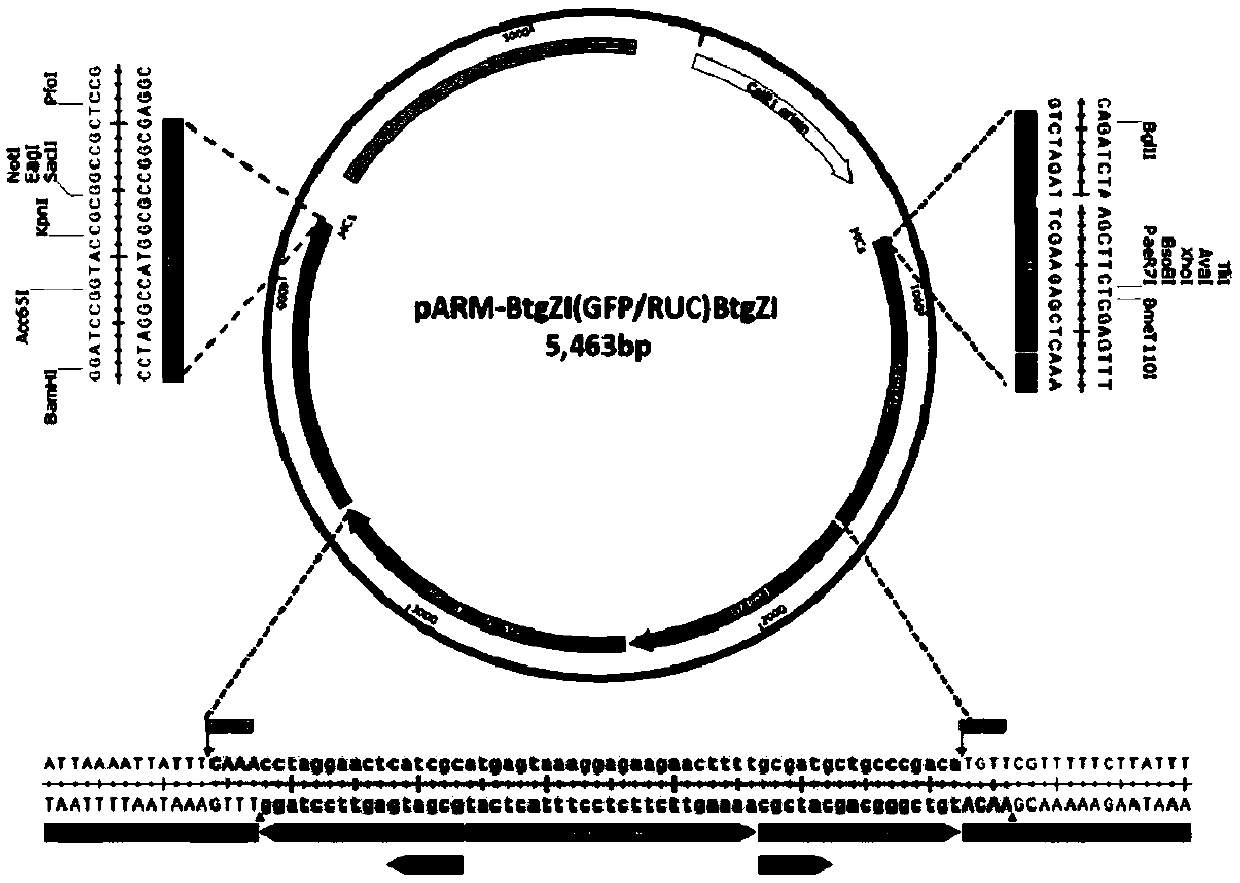
[0117] Example 2 Design and Construction of Universal Rescue Vector pARM-BtgZI(GFP / RUC)BtgZI
[0118] The structure of the rescue vector is as figure 2 As shown, its main components are: a 5' end promoter (5'PfEf1α, elongation factor 1-alpha, Gene ID: PF3D7_1357000) of the translation elongation factor of P. falciparum and Plasmodium berghei bifunctional Reporter gene GFP and renilla luciferase expression cassette (GFP / RUC), with BtgZI restriction sites at both ends of the GFP and renilla luciferase coding sequences, the reporter gene GFP / RUC can be replaced with other genes by cutting with BtgZI. There are multiple enzyme cutting sites (MCs) upstream of 5'-PfEf1α and downstream of 3'-PbDT for inserting 5' and 3' homology arms; an ampicillin (Amp) expression cassette, Used to screen positive clones and maintain the stability of the plasmid in Escherichia coli (such as DH5α, XL-10, Stbl3 and NEB Stable and other plasmid DNA cloning strains); The plasmid DNA replication ori...
Embodiment 3
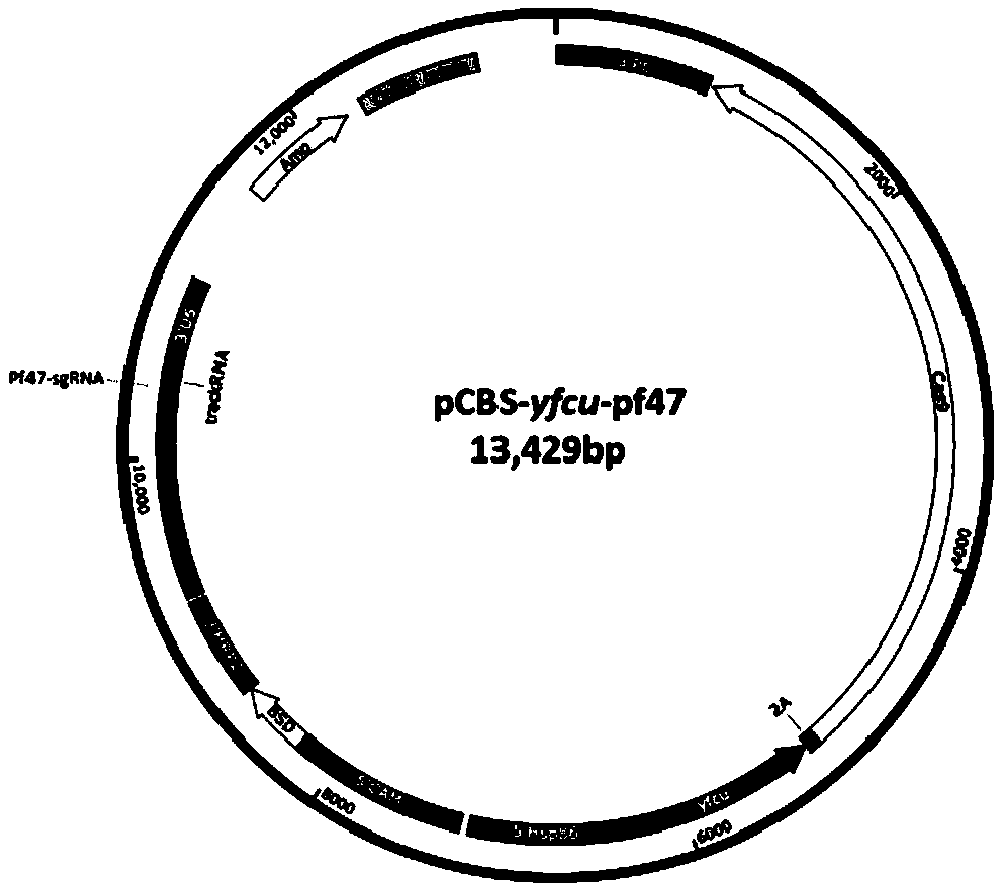
[0146] Example 3 Screening of Cas9 / sgRNA targets
[0147] Use sgRNAcas9 software (Java version) for target screening, import the target gene sequence and Pf genome sequence as required, and set parameters; the target sequence, that is, the sgRNA sequence should be 5'-N(19)GG, 5'-N( 20) GG or 5'-N(21)GG sequence, preferably 5'-N(20)GG sequence; since the present invention does not use in vitro transcription, but only constructs a common plasmid vector, the sgRNA sequence of the present invention refers to sgRNA Corresponding DNA sequence.
[0148] Select the Pf non-essential gene Pf47 as the target gene, enter the Pf47 gene sequence of Pf3D7 (GeneID: PF3D7_1346800) and the Pf3D7 genome sequence (downloaded from the PlasmoDB database) in the sgRNAcas9 software, and select the possibility of off-target in the genome according to the results given by the software The lowest target, its sequence is AACTACAGTTGGCTTAACATGG; in order to avoid the constructed vector being cut by Cas9,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com