Method for extracting trace agkistrodon halys venom genes, specific detection method and kit
An extraction method and specific technology, applied in the field of trace gene extraction from snake venom, can solve problems such as no specific primers and methods, difficulty in identification by non-professionals, missed treatment opportunities, etc., to achieve rapid identification, specific identification, and high sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Design of Viper gene-specific primers
[0055] According to the nucleotide sequence of Agkistrodon halys gene in the Gen Bank database, the online software (http: / / www.ncbi.nlm.nih.gov) was used for homology analysis and comparison, and the primer design software was used to determine the composition and composition of the base sequence. Analyze stability and other aspects, and refer to the principles of primer design, and design multiple pairs of primers with comprehensive consideration. The sequences of the primer pairs are as follows:
[0056]
Embodiment 2
[0057] Embodiment 2: Minor Genomic DNA Extraction
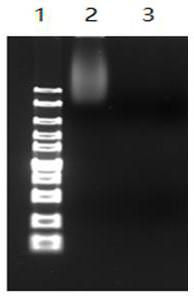
[0058] Take an appropriate amount of Agkistrodon halys tissue or toxin, put it in a centrifuge tube, add an appropriate amount of pure water with a final concentration of 100 μg / ml PK, incubate at 37°C, centrifuge for 10 minutes, suck the supernatant into another centrifuge tube, then add an equal volume of balanced phenol, Shake or invert to mix thoroughly, centrifuge at 12000g for 10min, absorb the supernatant water phase and place it in another centrifuge tube, add an equal volume of 1:1 mixed equilibrium phenol: chloroform, then shake or invert thoroughly, centrifuge at 12000g for 10min, absorb the supernatant water phase Place in another centrifuge tube, add an equal volume of chloroform, oscillate or invert to mix thoroughly, centrifuge at 12000g for 10min, absorb the supernatant water phase into another tube, add 2.5 times the volume of cold absolute ethanol to freeze for more than 10min, centrifuge at 12000g for 10min,...
Embodiment 3
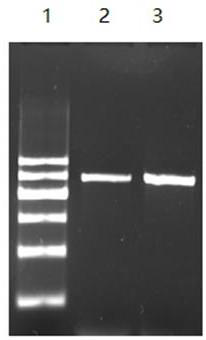
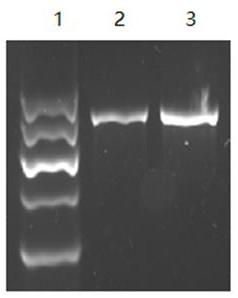
[0060] Embodiment 3: PCR amplification reaction
[0061] 1. PCR reaction conditions
[0062] The PCR reaction system includes: 20 μl total volume, 2.0 μl 10X PCR buffer; 2.0 μl 10X dNTPs, 0.2-1.5 μl upstream primer (final concentration: 0.2 μmol / l-1.5 μmol / l); 0.2-1.5 μl downstream primer (final concentration: 0.2μmol / l-1.5μmol / l); 0.4μl-2μl Taq enzyme (final concentration: 0.1 U / μl–0.5U / μl); DNA template 0.5μl; double distilled water to make up 20μl.
[0063] The PCR reaction parameters are:
[0064]
[0065] 2. Optimization of PCR reaction parameters
[0066] The preferred PCR reaction system is: total volume 20 μl, 10X PCR buffer 2.0 μl; 10X dNTPs 2.0 μl, upstream primer final concentration 0.5 μmol / l; downstream primer final concentration 0.5 μmol / l, Taq enzyme final concentration according to the reagent instruction manual; DNA Template 0.5μl; make up to 20μl with double distilled water.
[0067] The preferred PCR reaction temperature and time for primer pair 1 are...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com