A deep sequencing-based method suitable for the detection of HIV-1 protease inferior drug-resistant strains
An HIV-1PR, virus strain technology, applied in the biological field, can solve the problems of high detection cost, increased virological inhibition failure, single detection site, etc., and achieve the effect of reducing detection cost and sequencing cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0087] Example 1. Establishment of a deep sequencing method suitable for HIV-1PR drug resistance detection
[0088] 1. Primer combination
[0089] (1) Preparation of primers for the first round of RT-PCR
[0090] Take the synthesized primers DR1-1-CNB, DR2-1-CNBC, PRTM-F1a, PRTM-F1b, DR1-2-CNB and DR2-2-CNBC (Table 4), use ddH 2 O was sequentially diluted to a concentration of 20 μM. Mix the diluted primers, wherein DR1-1-CNB, DR2-1-CNBC, PRTM-F1a, PRTM-F1b are mixed according to 1 / 8 of the total volume, DR1-2-CNB, DR2-2-CNBC Mix them according to 1 / 4 of the total volume, mix well after mixing, and name the primer combination-IV.
[0091] (2) The second round of detection is intended to be used for the preparation of primers for deep sequencing
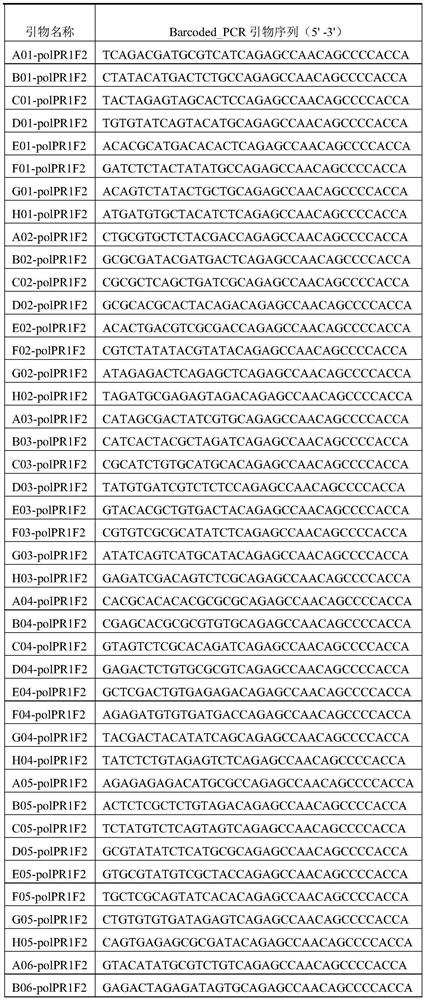
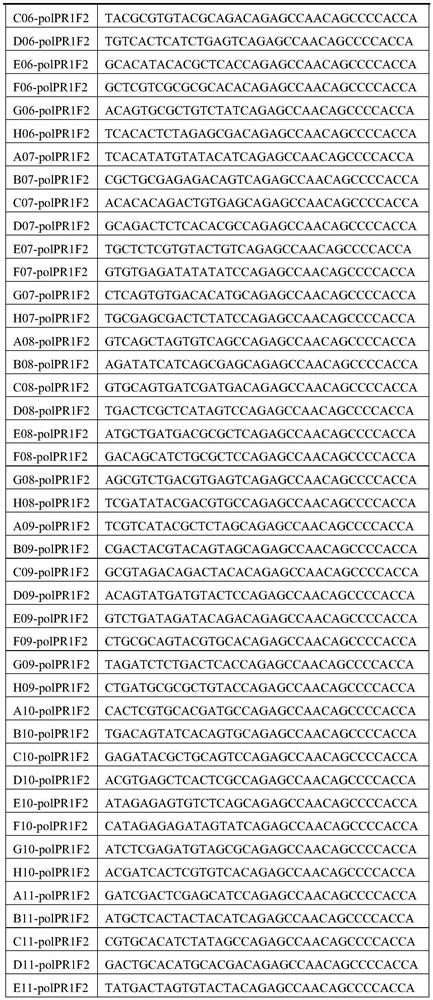
[0092] Use ddH for the primers in Table 2 and Table 3 2 Dilute O to a solution with a concentration of 20 μM, mix the primers in Table 2 and Table 3 in equal volumes according to the number of A01, B01, C01... in the primer name,...
Embodiment 2
[0102] Example 2. Sensitivity evaluation of deep sequencing method for HIV-1PR drug resistance detection
[0103] 1. Experimental samples
[0104] Plasma samples from 72 newly diagnosed HIV-infected patients who were about to receive antiretroviral therapy (all signed informed consent forms) were used as experimental samples, and the viral load of the infected patients was ≥1000Compies / ml.
[0105] 2. Using the established deep sequencing method to detect the drug resistance of HIV-1 infected PR region genes
[0106] The following experiments were performed on the plasma of each patient in step 1:
[0107] 1. Nucleic acid purification of plasma samples to be tested
[0108] All steps were performed according to the instructions of the nucleic acid purification kit MagNA Pure LC Total Nucleic AcidIsolation Kit (Code No. 3038505001) of Roche Company, and will not be repeated here.
[0109] 2. Preparation of deep sequencing samples
[0110] (1) Using the total RNA of step 1 a...
Embodiment 3
[0123] Embodiment 3, detection system specificity detection
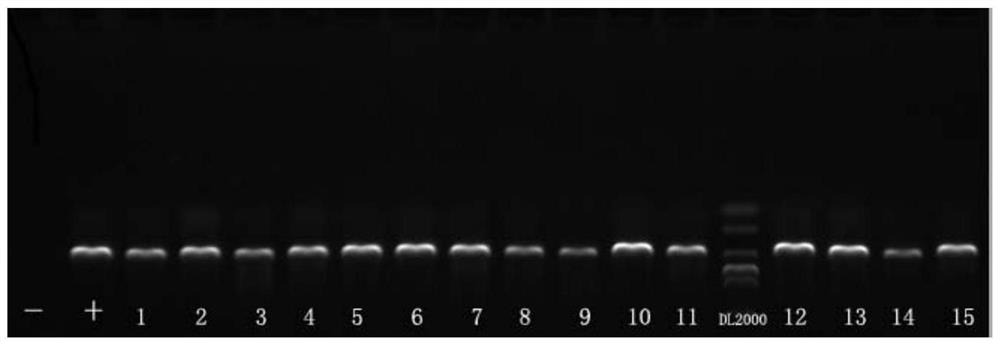
[0124] In order to verify whether the established detection system has cross-positive detection for similar viruses, HBV samples (representing human hepatitis B virus, 26 samples) and HCV (representing human hepatitis C virus, 40 samples) stored in the laboratory were taken. The testing process tests the sample. The detection of HCV samples completely followed the above detection reaction system and procedures; HBV reverse transcription at 50°C for 32 min was omitted in the first round of detection, and the detection system and procedures were the same as before. The test results show that the positive control CNHN24 test results have good specificity, HBV, HCV test samples and negative control (with ddH 2 O is the template) no specific target band appears, that is, the detection result is negative. The specific detection results confirmed that the established detection system had no cross-reaction to similar viru...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com