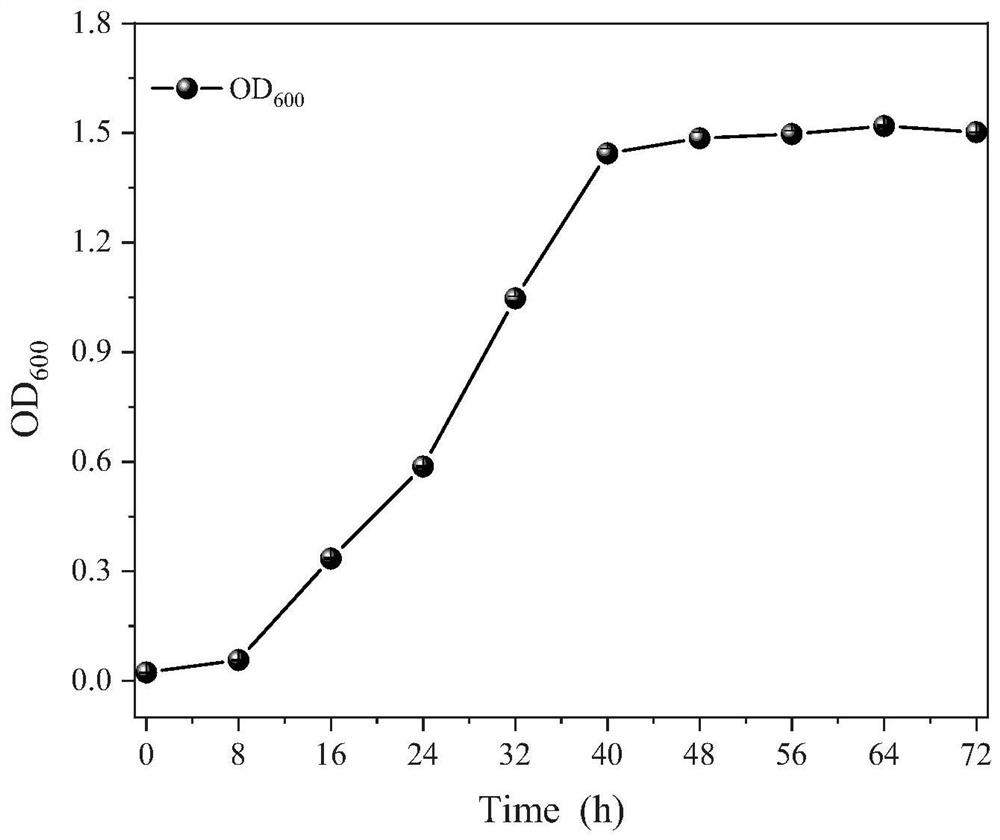
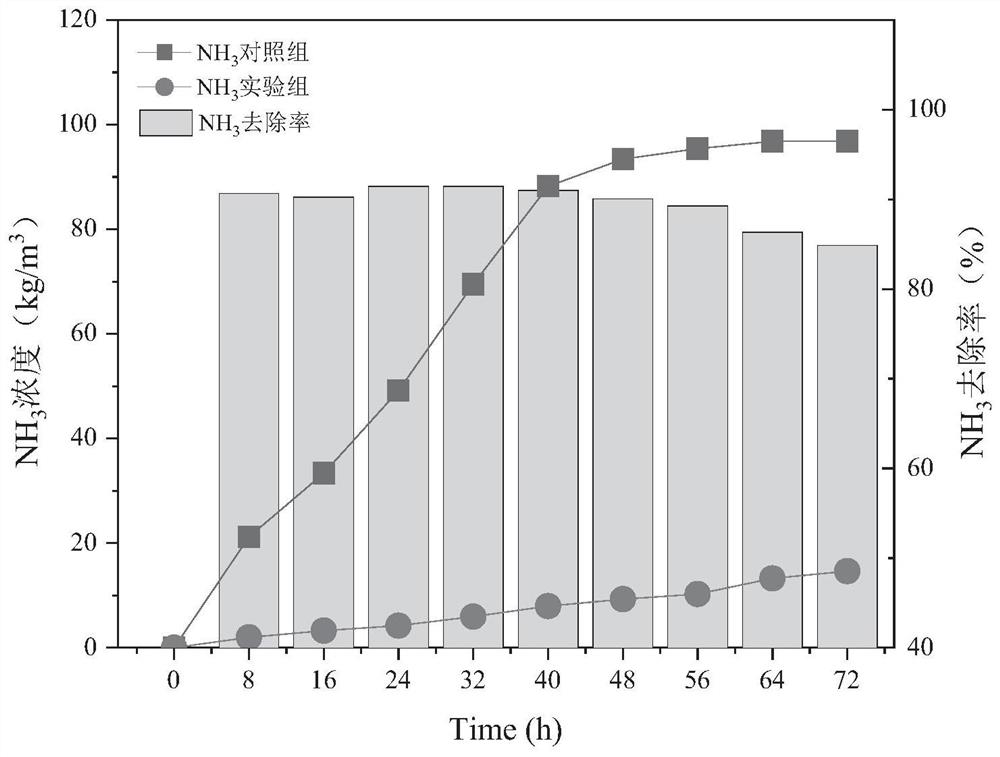
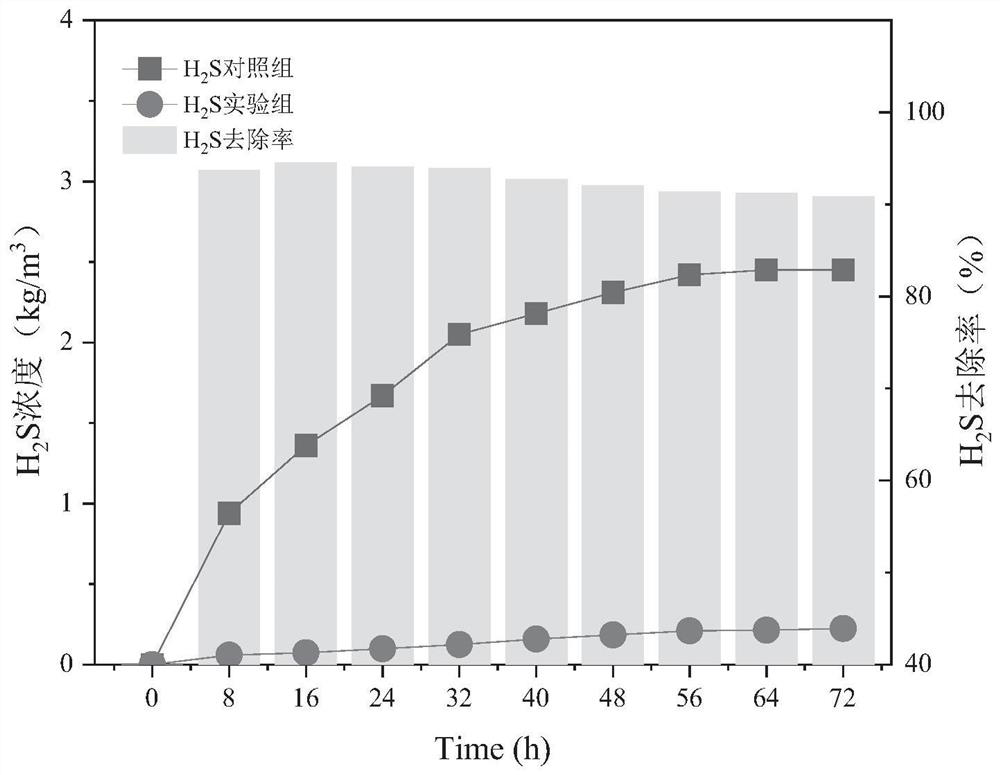
Efficient deodorizing bacteria and application thereof
A deodorizing bacteria, high-efficiency technology, applied in the direction of bacteria, special treatment targets, biochemical equipment and methods, etc., can solve the problems of high modification cost, inability to large-scale application, low microbial activity and adaptability, and achieve strong deodorization. The effect of odor efficacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Screening and Physiological and Biochemical Identification of Deodorizing Bacteria
[0025] (1) Source of strain
[0026] The bacteria source used in the present invention comes from solid waste in landfills.
[0027] (2) Isolation of strains
[0028] Take 10g of solid waste from a landfill, put it into a triangular flask containing sterilized glass beads and 100mL of normal saline, shake it at 30°C and 160r / min for 2h, let it stand for 20min, take 1mL of the supernatant, and incinerate The bacterial physiological water was diluted to a concentration of 10 -1 The suspension was serially diluted 10 times, and 200 μL of the suspension with different dilutions was drawn, spread on the medium, and cultured at 30°C for 28-72 hours. The growth of the strain was observed, and a single colony was picked for culture. According to the shape and growth of the strains, the strains with different shapes were picked for streak culture and stored at low temperature.
[...
Embodiment 2
[0041] Embodiment 2: the species identification of bacterial strain HCSW-2
[0042]By amplifying the 16S rDNA of the strain, a 16S rDNA sequence with a length of 1527 bp was obtained, as shown in SEQ ID NO.1. As PCR primers, bacterial universal primers 7F 5'-CAGAGTTTGATCCTGGCT-3' and 1540R 5'-AGGAGGTGATCCAGCCGCA-3' were used. The amplification reaction was carried out with a PCR machine.
[0043] PCR reaction system (25 μL): 2.5 μL 10×PCR Buffer, 1 μL dNTPs (concentration: 2.5 mmol / L), 0.5 μL each of the two primers, 0.5 μL DNA template, 0.5 μL rTaq DNA polymerase (5 U / μL), add Double distilled water to 25 μL.
[0044] The PCR amplification program was: 94°C pre-denaturation for 4 min, 94°C denaturation for 45 s, 55°C renaturation for 45 s, 72°C extension for 1 min, a total of 30 cycles, and finally 72°C extension for 10 min, 1% agarose gel electrophoresis, and EB staining UV detection. The homology comparison was carried out by the program, and the result showed that the ...
Embodiment 3
[0045] Example 3: Preparation of Lactobacillus plantarum HCSW-2 dormant cells
[0046] Preparation of dormant cells of strain Lactobacillus plantarum HCSW-2: Centrifuge the strain Lactobacillus plantarum HCSW-2 cultured in the medium in Example 1 at 8000r / min, 4°C for 10min, wash 3 times with phosphate buffer, and use the same Resuspend the precipitate in the buffer solution, so that the concentration of dormant cells is 10 mg / L, and freeze it at 4 ° C to obtain the dormant cells of the bacterial strain Lactobacillus plantarum HCSW-2, and prepare them for use. The method of activation within 6 months is the method of Example 1 culture medium for 36 hours at 36°C.
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| width | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com